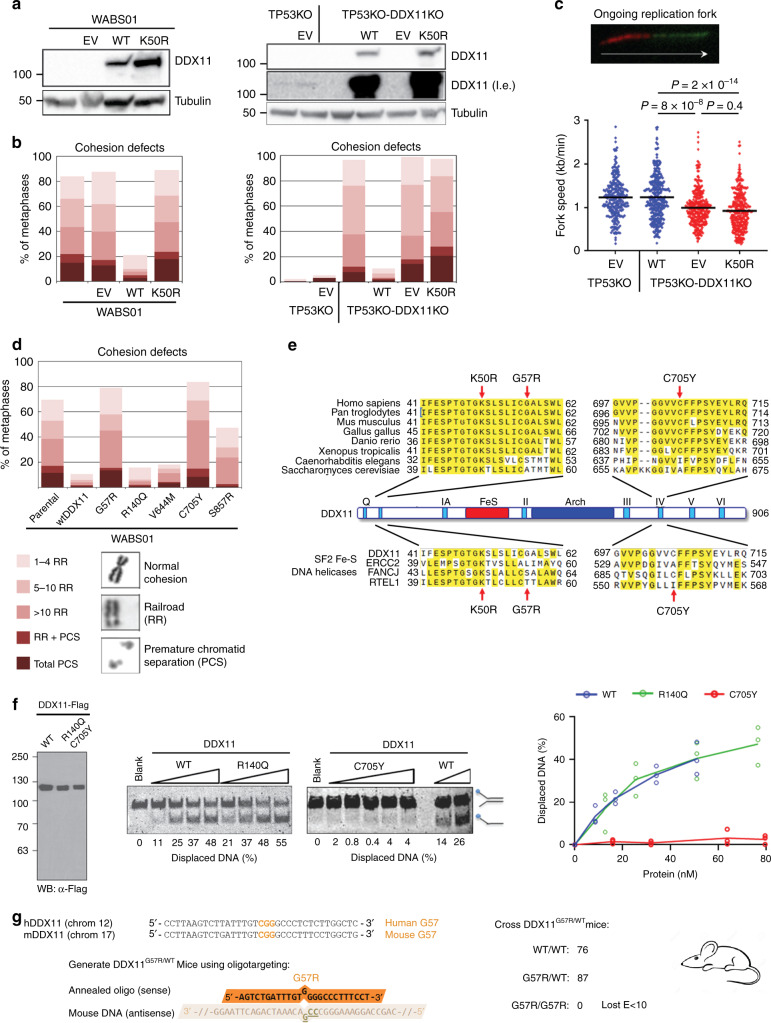
Fig. 8. DDX11 helicase activity supports cohesion, DNA replication and embryonic survival.
a WABS01 cells (left panel) and RPE1-TP53-DDX11KO cells (right panel) were stably transduced with empty vector (EV), wtDDX11 or DDX11-K50R and analyzed by Western blot. l.e. long exposure. A representative of two independent protein analyses is shown. b Cells were analyzed for cohesion defects. Per condition, in total 100 metaphases from two independent experiments were assessed. c Replication fork speed was assessed with a DNA fiber assay using a double labeling protocol. In total at least 200 fibers were scored per condition in two independent experiments. The example track represents an ongoing fork. Blue dots, DDX11 proficient; red dots, DDX11 deficient. Black lines indicate the mean. P-values were calculated by a non-parametric one-way ANOVA test. d WABS01 cells, stably transfected with different DDX11 mutants, were analyzed for cohesion defects. Per condition, in total 150 metaphases from three independent experiments were assessed. e Alignment of different DDX11 orthologues (top) and related SF2 DNA helicases (bottom). Conserved motifs are indicated. Arrows point at the positions of K50R, G57R, and C705Y mutations. f In vitro DNA helicase assay of patient-derived DDX11 mutants. Purified, recombinant Flag-tagged proteins were analyzed by western blot (left). Proteins were incubated at increasing concentrations with a fluorescently labeled forked DNA substrate. Helicase reaction products were resolved by gel electrophoresis. Percentage of displaced DNA was determined using Image-J software. Right panel: quantification from three independent experiments. g DDX11-G57R heterozygous mice were generated by oligonucleotide-directed gene modification in mouse embryonic stem cells substituting glycine codon 57 for an arginine codon (C>G substitution). Subsequent intercrossing of heterozygous mice failed to produce homozygous offspring.