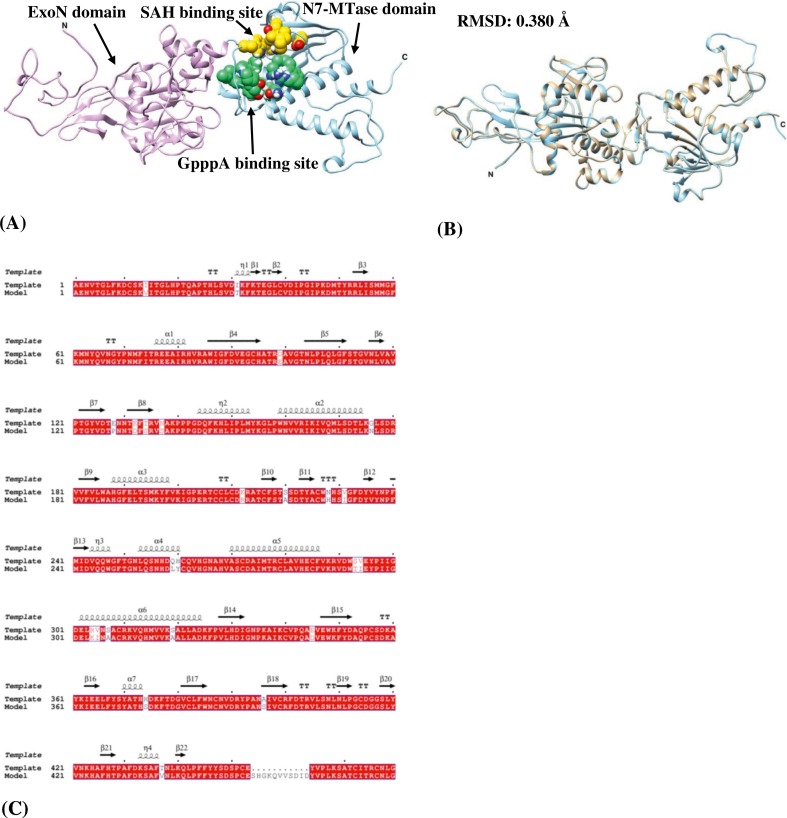
Fig. 2.
(A) In silico model structure of SARS-CoV-2 Nsp14 showing two domains in ribbons – ExoN domain: 1–287 residues (plum) and N7-MTase domain: 288–527 residues (sky blue). The ligand-binding pocket consists of SAH binding site (gold spheres) which encompasses residues-Trp292, Asp331, Gly333, Pro335, Lys336, Asp352 and Val389 and GpppA binding site (spring green spheres) contain residues such as Asn306, Arg310, Trp385, Asn386, Phe401, Leu419, Tyr420, Asn422, Lys423, Phe426, Thr428 and Phe506. (B) Superposition of the silico model structure of SARS-CoV-2 Nsp14 (cyan) with the template (tan) (C) pairwise sequence alignment between SARS-CoV-2 Nsp14 and the template protein with identical residues represented by white characters inside the red box. The secondary elements such as α-helices and 310-helices (η) are rendered as coils whereas β-strands, strict β-turns and strict α-turns are represented by arrows, TT letters and TTT letters respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)