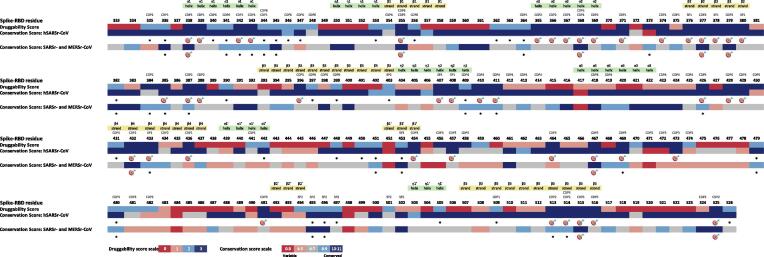
Fig. 2.
Overall alignment of Spike-RBD druggability along with the conservation scores for each residue position. The druggability prediction was based on the descriptors algorithm of each pocket bioinformatics tool: SF, DGSS and PDS. The potential conserved druggable sites/residues are marked with an asterisk and the T-RHS are marked with a target. The conserved druggable pockets allocated to each site/residue along with the secondary structure elements are indicated at the top of the picture.