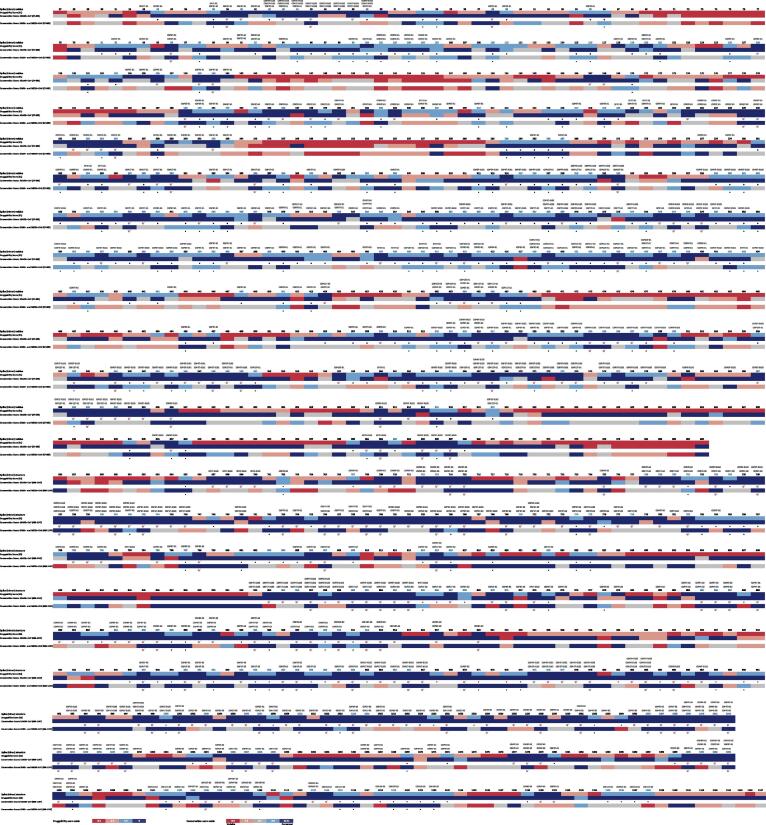
Fig. 5.
Overall alignment of Spike S1 and S2 druggability along with the conservation scores for each residue position. The druggability prediction was based on the descriptors algorithm of each pocket bioinformatics tool: SF and DGSS; and for all the predicted S state conformation (open, semi-open and open). The potential conserved druggable sites/residues are marked with an asterisk and the T-RHS are marked with a target. The conserved druggable residues shared by the S monomer and trimer conformations for the hSARSr-CoVs are coloured in blue; and the conserved druggable pockets allocated to each site/residue are indicated at the top of the picture. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)