Abstract
Juvenile neuronal ceroid lipofuscinosis (JNCL, aka. juvenile Batten disease or CLN3 disease), a lethal pediatric neurodegenerative disease without cure, often presents with vision impairment and characteristic ophthalmoscopic features including focal areas of hyper-autofluorescence. In the associated research article “Loss of CLN3, the gene mutated in juvenile neuronal ceroid lipofuscinosis, leads to metabolic impairment and autophagy induction in retinal pigment epithelium” (Zhong et al., 2020) [1], we reported ophthalmoscopic observations of focal autofluorescent lesions or puncta in the Cln3Δex7/8 mouse retina at as young as 8 month old. In this data article, we performed differential interference contrast and confocal imaging analyses in all retinal layers to localize and characterize these autofluorescent lesions, including their spectral characteristics and morphology. We further studied colocalization of these autofluorescent lesions with the JNCL marker mitochondrial ATP synthase F0 sub-complex subunit C and various established retinal cell type markers.
Keywords: Juvenile Neuronal Ceroid Lipofuscinosis, CLN3, vision loss, retinopathy, autofluorescent lesions, mitochondrial ATP synthase F0 sub-complex subunit C
Specifications table
| Subject | Biochemistry, Genetics and Molecular Biology (General) |
| Specific subject area | Phenotypical characterization of neurodegenerative disease |
| Type of data | Image, figure |
| How data were acquired | Murine model, immunohistochemistry, fluorescent staining, confocal microscopy, differential interference microscopy |
| Data format | Raw, analysed |
| Parameters for data collection | Cln3Δex7/8 and wild type control mouse retina, young vs. old mice. Additional controls include retinal sections processed without either primary antibody or fluorescently labelled peanut agglutinin. |
| Description of data collection | Two-color confocal images were acquired in serial mode and using identical instrument settings such as laser power, detector gain, pixel dwell time, image size, pinhole size, method of average/summation, and scan zoom within each sample set. To make figures, images from the same sample set were processed identically for comparing different samples within the set. |
| Data source location | University of Kentucky, Lexington, Kentucky, United States Latitude and longitude (and GPS coordinates) for collected samples/data: 38.0406° N, 84.5037° W |
| Data accessibility | With this Data in Brief article and raw data deposited in Mendeley Data (http://doi.org/10.17632/gwhbbtkpx6) |
| Related research article | Zhong, Y., Mohan, K., Liu, J., Al-Attar, A., Lin, P., Flight, R.M., Sun, Q., Warmoes, M.O., Deshpande, R.R., Liu, H., Jung, K.S., Mitov, M.I., Lin, N., Butterfield, D.A., Lu, S., Liu, J., Moseley, H.N.B., Fan, T.W. M., Kleinman, M.E., Wang, Q.J. (2020). Loss of CLN3, the gene mutated in juvenile neuronal ceroid lipofuscinosis disease, leads to metabolic impairment and autophagy induction in retinal pigment epithelium. BBA-Mol Basis Dis. DOI: 10.1016/j.bbadis.2020.165883. |
Value of the data
Despite frequent observation of autofluorescent lesions in the JNCL retina, these lesions are largely uncharacterized. The imaging data reported in this data article provide a detailed characterization of the retinal autofluorescent lesions in a JNCL mouse model.
These data, which reveal specific features of the JNCL retinal autofluorescent lesions, including their spectral characteristics, morphology, and potential colocalization with JNCL marker mitochondrial ATP synthase F0 sub-complex subunit C and various established retinal cell type markers, may provide useful information for researchers and clinicians who seek better understanding and treatment of JNCL disease.
These data may stimulate further investigations that identify and compare retinal cell type(s) that harbor the retinal autofluorescent lesions in JNCL patients and animal models. This knowledge not only may give insights into potential etiology of JNCL vision loss, but also may provide a potential biomarker for evaluating disease progression and therapeutic efficacy.
1. Data description
In this data article, we first performed confocal fluorescent microscopy on the retinal cryo-sections from young (6 month-old) and old (22 month-old) homozygous Cln3Δex7/8 mice to visualize the distribution of the pathological autofluorescent lesions across all retina layers (Fig. 1A). Second, we analyzed the spectral properties of these retinal autofluorescent lesions in old homozygous Cln3Δex7/8 mice using spectral imaging (Fig. 1B). Next, we imaged morphology of these retinal autofluorescent lesions using differential interference contrast microscopy (DIC, Fig. 2, Fig. 3, Fig. 4, Fig. 5, top panels) and accessed the colocalization of these retinal autofluorescent lesions with mitochondrial ATP synthase F0 sub-complex subunit C (abbreviated subunit C hereafter), a JNCL lysosomal aggregate marker, using immunohistochemistry-immunofluorescence (IHC-IF) and confocal fluorescent microscopy (Fig. 2). Finally, we also evaluated the colocalization of these retinal autofluorescent lesions with various established retinal cell type markers, including peanut agglutinin (PNA, a cone photoreceptor outer segment, inner segment, and pedicle marker; Fig. 3), ionized calcium binding adaptor molecule 1 (IBA1, a microglial cell marker; Fig. 4) and CHX10/Visual System Homeobox 2 (VSX2) (a bipolar cell nuclear marker; Fig. 5). Raw data (i.e., .nd2 files that contain information of all image acquisition settings) are listed in the Supplemental Table and deposited in Mendeley Data (http://doi.org/10.17632/gwhbbtkpx6).
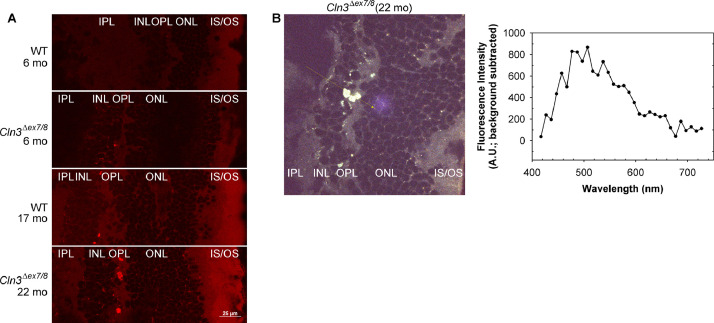
Fig. 1.
Homozygous Cln3Δex7/8 mouse retina showed pathological autofluorescent lesions predominantly in the retinal outer plexiform and inner nuclear layers.
(A) Confocal fluorescent images of mouse retinal cryo-sections show that autofluorescent lesions, which were prominently in the outer plexiform and inner nuclear layers, increased in size and number with age and homozygous Cln3Δex7/8 mutation. Confocal images were collected with 561.4 nm laser and 595/50 nm emission filter on a Nikon A1plus confocal microscope. These fluorescent microscope settings were similar to the 536/40 nm excitation filter and 650/100 nm barrier filter settings that we used to first observe focal autofluorescent lesions in the Cln3Δex7/8 mouse retina by fundus autofluorescence (FAF) imaging on a TRC 50-IX imaging system [1]. Of note, we observed weak autofluorescence from RPE or choroid (see the raw image files for Fig. 1A in Mendeley Data), likely due to extensive light absorption by the pigments in the melanosomes in these retinal layers [6]. In addition, the confocal fluorescent images of mouse retinal cryo-sections collected using either 403.1 nm laser and 450/50 nm emission filter or 487.9 nm laser and 525/50 nm emission filter show similar autofluorescence patterns to those collected using 561.4 nm laser and 595/50 nm emission filter (see the raw image files for Fig. 1A in Mendeley Data). Scale bar: 25 μm. Labels: OS, outer segment; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer. (B) Spectral imaging showed that autofluorescent lesions in homozygous Cln3Δex7/8 retina had a broad spectrum peaked at ∼500 nm. Left panel: An autofluorescent image of a 22-month old homozygous Cln3Δex7/8 mouse retinal cryo-section combined from all 32 channels of the spectral detector (centered at 417 nm, 427 nm, …, and 727 nm). Right panel: The light spectrum of the autofluorescent lesions seen in the left panel. To obtain the spectrum from the images, moving averages (n=6) of signals were calculated along the line of interest (yellow line with arrow) for each of the 32 channels. Differences between the averaged signals within the autofluorescent lesions and at a neighboring background area were calculated and plotted against the corresponding center wavelengths of the channels.
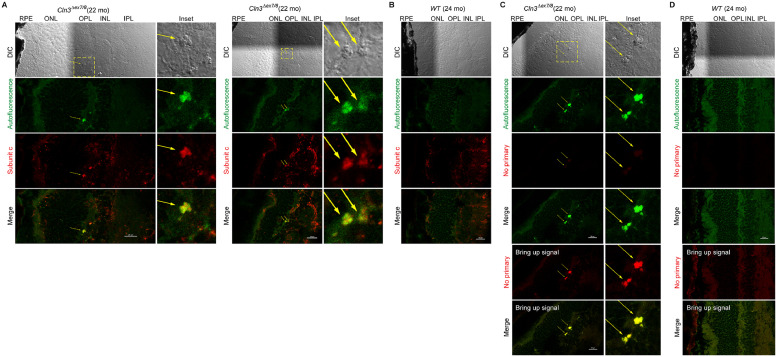
Fig. 2.
Autofluorescent lesions in the homozygous Cln3Δex7/8 mouse retinal OPL/INL were immunoreactive for mitochondrial ATP synthase F0 sub-complex subunit C.
Immunoreactivity of subunit C, a marker known to accumulate in the lysosomes and ceroids/lipofuscins in JNCL [7, 8], was previously reported in the ONL, INL, and RGC of 10 month old homozygous Cln3Δex7/8 mice on an outbred 129Sv/Ev/CD1 mixed background [2]. However, CD1 background itself carried retinal degenerative genetic loci [9]. Here we observed subunit C immunoreactivity in the photoreceptor IS, OPL, INL and IPL in both WT and homozygous Cln3Δex7/8 mice (22-24 month old) on a C57BL/6J background which did not harbor the retinal degenerative Rd1 or Rd8 mutation. Specifically, autofluorescent lesions (green) prominent in the OPL and INL of the homozygous Cln3Δex7/8 mouse retina was subunit C immunoreactive. Due to the spectral properties of the autofluorescent lesions in the homozygous Cln3Δex7/8 mouse retina (see Fig. 1), autofluorescence was collected with a 487.9 nm laser and a 525/25 nm emission filter. Subunit C IHC-IF was performed on retinal cyro-sections stained with a rabbit polyclonal anti-subunit C antibody (Abcam ab181243) followed by an Alexa Fluor Plus 647-labeled secondary antibody against rabbit IgG(H+L) (Thermo Fisher Scientific A32733) and confocal fluorescent images were acquired with a 638.6 nm laser and a 700/75 nm emission filter. (A) Representative DIC and confocal fluorescent images of homozygous Cln3Δex7/8 mouse retina show autofluorescent lesions (green channel; pointed by yellow arrows) in the OPL/INL immunoreactive for subunit C (red, pseudo-color). (B) Representative DIC and confocal fluorescent images of WT mouse retina show lack of autofluorescent lesions (green) in the OPL/INL despite subunit C immunoreactivity in other areas (red, pseudo-color). (C) Control fluorescent images of homozygous Cln3Δex7/8 mouse retina acquired and processed exactly as in (A) except that anti-subunit C was omitted during immunostaining. Note that bringing up signals in the red channel revealed autofluorescence of the autofluorescent lesions rather than subunit C immunoreactivity in the OPL/INL. (D) Control fluorescent images of WT mouse retina acquired and processed exactly as in (B) except that anti-subunit C was omitted during immunostaining.
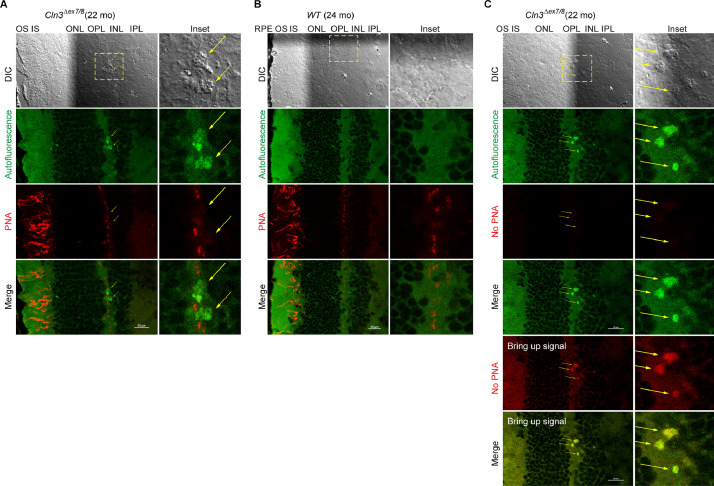
Fig. 3.
Autofluorescent lesions in the homozygous Cln3Δex7/8 mouse retinal OPL/INL were negative for the cone photoreceptor marker peanut agglutinin.
Peanut agglutinin (PNA), a lectin protein derived from the fruits of Arachis hypogaea and with high affinity for galactose-galactosamine disaccharide residues, stains cone photoreceptors [10,11]. Hypopigmented homozygous Cln3Δex7/8 mice (10-17 month old) on an outbred 129Sv/Ev/CD1 mixed background, which is known to harbor additional CLN3-independent retinal degenerative genetic loci, were shown to have significantly decreased cone photoreceptors by PNA chromogenic IHC [2,9]. However, we did not observe overt photoreceptor loss by Cy5-PNA fluorescence in a homozygous Cln3Δex7/8 mouse (22 month old) on a C57BL/6J background. In addition, autofluorescent lesions in the homozygous Cln3Δex7/8 mouse retinal OPL/INL were negative for Cy5-PNA. Autofluorescence was collected with a 487.9 nm laser and a 525/25 nm emission filter. PNA IF was performed on retinal cyro-sections stained with Cy5-conjugated PNA (Vector CL-1075a) and confocal fluorescent images were acquired with a 638.6 nm laser and a 700/75 nm emission filter. (A) Representative DIC and confocal fluorescent images of homozygous Cln3Δex7/8 mouse retina show autofluorescent lesions in the OPL/INL (green channel; pointed by yellow arrows) and Cy5-PNA fluorescence (red; pseudo-color) at the cone IS and OS and cone pedicles in the OPL/INL. Note that DIC images clearly show distinctive morphologies of RPE, OS, IS, ONL, OPL, INL, IPL, and autofluorescent lesions. (B) Representative DIC and confocal fluorescent images of WT mouse retina show autofluorescence (green) and Cy5-PNA fluorescence (red; pseudo-color). (C) Control fluorescent images of homozygous Cln3Δex7/8 mouse retina acquired and processed exactly as in (A) except that Cy5-PNA staining was omitted. Note that bringing up signals in the pseudo-red channel revealed autofluorescent lesions rather than the typical Cy5-PNA staining patterns (A-B) in the OPL/INL.
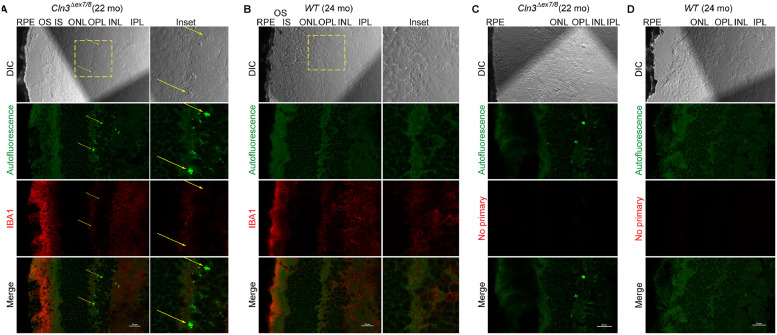
Fig. 4.
Autofluorescent lesions in the homozygous Cln3Δex7/8 mouse retinal OPL/INL were negative for immunostaining of IBA1.
IBA1 was reported as a marker for microglial cells and their activation in the retina, including photoreceptor IS and OS, OPL, IPL and RGC layers [12]. Here we observed IBA1 immunoreactivity primarily in the photoreceptor OS layer for both homozygous Cln3Δex7/8 and WT mouse retina. Autofluorescence lesions in the homozygous Cln3Δex7/8 mouse retinal OPL/INL did not show IBA1 IHC-IF signal beyond autofluorescence seen in the absence of IBA1 antibody. (A) Representative DIC and confocal fluorescent images of homozygous Cln3Δex7/8 mouse retina show autofluorescent lesions (green channel; pointed by yellow arrows) in the OPL/INL and immunoreactivity of IBA1 (red; pseudo-color). Autofluorescence was collected with a 487.9 nm laser and a 525/25 nm emission filter. IBA1 IHC-IF was performed on retinal cyro-sections stained with a rabbit polyclonal anti-IBA1 antibody (Fujifilm 019-19741; distributed as VWR 100369-764) followed by an Alexa Fluor Plus 647-labeled secondary antibody against rabbit IgG(H+L) (Thermo Fisher Scientific A32733) and confocal fluorescent images were acquired with a 638.6 nm laser and a 700/75 nm emission filter. (B) Representative DIC and confocal fluorescent images of WT mouse retina show autofluorescence (green) and immunoreactivity for IBA1 (red, pseudo-color). (C) Control fluorescent images of homozygous Cln3Δex7/8 mouse retina acquired and processed exactly as in (A) except that anti-IBA1 was omitted during immunostaining. (D) Control fluorescent images of WT mouse retina acquired and processed exactly as in (B) except that anti-IBA1 was omitted during immunostaining.
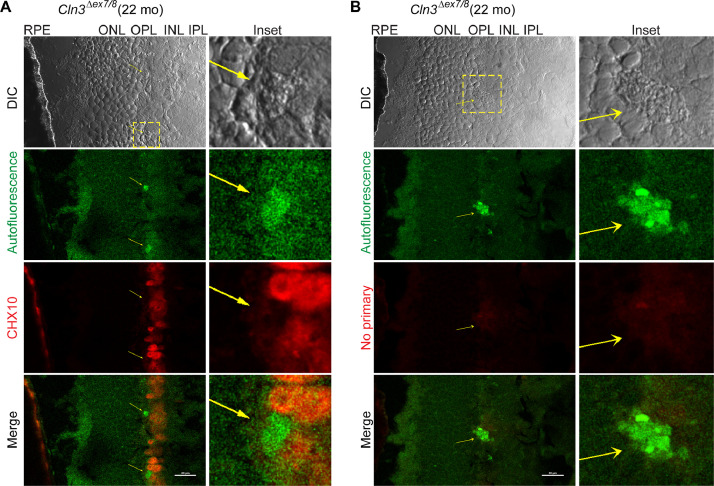
Fig. 5.
Autofluorescent lesions in the homozygous Cln3Δex7/8 mouse retinal OPL/INL were negative for immunostaining of the bipolar cell nuclear marker CHX10.
CHX10/VSX2 is a homeobox-containing transcription factor required for retinal progenitor cell proliferation and bipolar neuron differentiation [13,14]. Here we observed autofluorescent lesions in the homozygous Cln3Δex7/8 mouse retinal OPL/INL next to CHX10-positive bipolar cell nuclei. This observation warrants future testing of potential colocalization of autofluorescent lesions with a cytosolic bipolar cell marker, such as protein kinase C α isoform (PKCα) for rod bipolar cells [15], [16], [17] and secretagogin for cone bipolar cells [17]. As 15 types of bipolar cells have been classified so far [18], autofluorescent lesions may colocalize with certain classes of bipolar cells in the homozygous Cln3Δex7/8 mouse retina. (A) Representative DIC and confocal fluorescent images of homozygous Cln3Δex7/8 mouse retina show bipolar cell nuclei immunostained by an anti-CHX10 antibody [19] (red; pseudo-color) and autofluorescent lesions (green channel; pointed by yellow arrows) in the OPL/INL. Autofluorescence was collected with a 487.9 nm laser and a 525/25 nm emission filter. CHX10 IHC-IF was performed on retinal cyro-sections stained with a mouse monoclonal anti-CHX10 IgG2a antibody (Santa Cruz Biotechnology sc-365519) followed by an Alexa Fluor 680-labeled secondary antibody against mouse IgG(H+L) (Thermo Fisher Scientific A21058) and confocal fluorescent images were acquired with a 638.6 nm laser and a 700/75 nm emission filter. (B) Control fluorescent images of homozygous Cln3Δex7/8 mouse retina acquired and processed exactly as in (A) except that anti-CHX10 was omitted during immunostaining.
2. Experimental design, materials, and methods
2.1. Mouse strains and genotyping
C57BL/6J wild-type (WT; 000664) and JNCL mutant B6.129(Cg)-Cln3tm1.1Mem/J (Cln3Δex7/8; 017895; made by Cotman et al. [2] and backcrossed to C57BL/6J) mice were purchased from the Jackson Laboratories (Bar Harbor, ME) and housed under standard conditions with a 14:10 h light-dark cycle. For genotyping WT and Cln3Δex7/8 mutant mice, genomic DNAs were prepared from mouse ear punches by overnight proteinase K digestion (100 μg per 350 μL lysis buffer) at 55°C, followed by isopropanol (70%) precipitation and ethanol (50%) wash. Air-dried genomic DNAs were resuspended in ddH2O and used as the templates for Polymerase Chain Reactions (PCRs), with 50-100 ng genomic DNAs per reaction. Using the WT forward (oIMR3012, 5’-CAC CTT CCT CTC ACT GAC TGC-3’) and reverse (oIMR3013, 5’-ACC ACC ATG AGA TCA CAG CA-3’) primers, WT mice showed a 145 bp band (including primer length) that the mutants do not have. Using the mutant forward (5’-GCC TTT ACT TGC TGC CTT ACA-3’) and reverse (5’-GGG TCT CGG TGC CTA TGA-3’) primers, the mutants showed a band at ∼750 bp band, while WT mice showed a fainter 2.7 kb band. PCRs were carried out using a Taq DNA Polymerase Kit (QIAGEN 201205) with the optimized annealing temperature at 56 °C.
We also genotyped both WT and Cln3Δex7/8 mutant strains (both on C57BL/6J background) for potential presence of the retinal degeneration Pde6b (Rd1) mutation [3] and the Crb1 (Rd8) mutation [4]. We did not find evidence for the presence of either mutation in these mice. For genotyping Rd8 or Rd1 mutation, genomic DNAs were prepared from mouse ear punches and PCRs were carried out as described above. Primers for genotyping the Rd1 mutation include oIMR2093 (5’-AAG CTA GCT GCA GTA ACG CCA TTT-3’) for the Rd1 mutation, oIMR2094 (5’-ACC TGC ATG TGA ACC CAG TAT TCT ATC-3’) for WT, and oIMR2095 (5’-CTA CAG CCC CTC TCC AAG GTT TAT AG-3’) as the common primer. With these primers, WT showed a 240 bp band while the mutant showed a band at 560 bp. The optimized PCR annealing temperature was 65°C.
The Rd8 mutation is a single-base deletion mutation c.3481delC in the Crb1 gene, i.e., WT sequence TTCTTATCGGTGTG and Rd8/Rd8 sequence TTCTTATGGTGTG. Primers for genotyping the Rd8 mutation were forward primer (5’-GGT GAC CAA TCT GTT GAC AAT CC-3’) and reverse primer (5’-GCC CCA TTT GCA CAC TGA TGA C-3’). The optimized PCR annealing temperature was 55°C. The PCR amplicons (∼434 bp) were resolved on a 1% agarose gel containing 0.01% Sybr™ Safe DNA Gel Stain (Thermo Fisher Scientific S33102), excised, and cleaned using a QIAquick gel purification kit (QIAGEN 28104). Rd8 mutation was tested using the above primers by Sanger sequencing in the University of Kentucky Chandler Hospital Genomics Core Laboratory. To carry out Sanger sequencing, cleaned amplicons (3 µl of ∼15 ng/µl) were sequenced in 10 µl reaction with 0.5 µl each of the forward and reverse primer stocks (5 µM).
2.2. Tissue preparations and microscopy
Mice were euthanized by CO2 asphyxiation (20% cage volume per minute) followed by cervical dislocation. Eyes were enucleated and cryo-preserved in Tissue-Tek O.C.T compound (Sakura, 4583). Cross sections (10 µm thick) of mouse eyes were cut on a cryostat and adhered to glass slides. For imaging retinal autofluorescence in Fig. 1, cryo-sections were fixed in 4% paraformaldehyde for 15 min, washed 3 times with 1 × PBS with 0.1% Tween-20, sealed in Vectashield mounting medium (Vector laboratories, H-1400) at 4 °C in the dark overnight. Regular confocal images of autofluorescence (Fig. 1A) were collected on a Nikon A1plus confocal microscope, with a Plan Apo λ 100x oil objective lens, lasers (403.1 nm, 487.9 nm and 561.4 nm) and corresponding emission filter cubes (450/50, 525/50 and 595/50 nm, respectively). Laser powers and detector gains were set the same for all samples. Z-stacks were acquired with the most intense plane in the red channel selected to be included in the figure. Spectral imaging of retinal autofluorescence (Fig. 1B) was acquired on the same Nikon A1plus confocal microscope, with a Plan Apo λ 100x oil objective lens, a 403.1 nm laser, and a spectral detector divided into 32 channels (centered at 417 nm, 427 nm, …, and 717 nm).
For Cy5-PNA staining (Fig. 3), retinal cryo-sections were air-dried for 90 min, fixed in 4% paraformaldehyde for 40 min, washed 2 times with 1 × PBS, stained with Cy5-PNA (Vector CL-1075; 10 µg/mL) for 1 h in 1 × PBS, washed 3 times with 1 × PBS, and sealed in ProLong® Diamond antifade reagent (Invitrogen P36961) and cured in the dark for at least 24 h. Control retinal cryo-sections were processed identically except for not adding Cy5-PNA during staining. For DIC, Köhler illumination was set up and images were acquired on a Nikon A1plus confocal microscope with an Apo 60x oil λS DIC N2 objective lens and a 487.9 nm laser. Autofluorescence was acquired with the same objective lens, same 487.9 nm laser, same laser power as those for DIC, and a 525/50 nm emission filter. Cy5-PNA fluorescence was acquired with a 638.6 nm laser and a 700/75 nm emission filter. Images were acquired in serial mode and using identical instrument settings (laser power, detector gain, pixel dwell time, image size, pinhole size, method of average/summation, and scan zoom) for all samples in one sample set. Imaging stitching (2 by 2) was used to include all retinal layers in one larger image. Z-stacks were acquired and the best-focused plane was selected to be included in the figure. Fluorescent images from the same sample set were processed identically post-acquisition.
For subunit C (Fig. 2) and IBA1 (Fig. 4) IHC-IF, retinal cryo-sections were air-dried for 90 min, fixed in 4% paraformaldehyde for 40 min, washed 2 times with 1 × PBS, permeabilized for 20 min with 0.1% Triton X-100 in 1 × PBS supplemented with 2% BSA 2% normal goat serum (NGS), blocked for 40 min in 1 × PBS supplemented with 2% BSA 2% NGS, incubated with corresponding primary antibodies anti-subunit C (Abcam ab181243, 1:200) and anti-IBA1 (Fujifilm 019-19741 and distributed as VWR 100369-764, 1:500), respectively, in 1 × PBS supplemented with 2% BSA 2% NGS at 4 °C overnight in a humidified chamber, washed 3 times with 1 × PBS, incubated with an Alexa Fluor Plus 647-labeled secondary antibody against rabbit IgG(H+L) (Thermo Fisher Scientific A32733, 1:1000) at room temperature for 1 h, washed 3 times with 1 × PBS, and sealed in ProLong® Diamond antifade reagent and cured in the dark for at least 24 h. Control retinal cryo-sections were processed identically except for not adding primary antibodies during immunostaining. For CHX10 (Fig. 5) IHC-IF, retinal cryo-sections were processed similarly except that permeabilization was done for 20 min with 0.2% Triton X-100 in 1 × PBS, and primary and secondary antibodies were anti-Chx10 (Santa Cruz Biotechnology sc-365519, 1:200) and Alexa Fluor 680-labeled secondary antibody against mouse IgG(H+L) (Thermo Fisher Scientific A21058), respectively. DIC, autofluorescence, and subunit C (Fig. 2), IBA1 (Fig. 4) and CHX10 (Fig. 5) IHC-IF images were acquired and processed as described above for Cy5-PNA (Fig. 3).
3. Note
While this manuscript was under review in Data in Brief, a publication came online reporting reduced number of bipolar cells in Cln3Δex7/8 mice and rescue of ocular phenotype by adeno-associated virus (AAV)-mediated, bipolar cell-specific expression of CLN3 in Cln3Δex7/8 mice [5], consistent with our finding of potential cytoplasmic localization of autofluorescent lesions in bipolar cells of Cln3Δex7/8 mice (Fig. 5).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
Acknowledgements
This work was supported by National Institutes of Health Center of Biomedical Research Excellence (COBRE) award P20GM121327 (pilot grant to Q.J.W.), and National Institutes of Health EY028206 and BrightFocus Foundation (to M.E.K.). Q.J.W. also thank Drs. Susan L. Cotman (Massachusetts General Hospital) and Wai T. Wong (National Eye Institute, NIH) for suggestions of subunit C and IBA1 antibodies, respectively .
Ethics statement
All animal experiments were approved by the University of Kentucky Institutional Animal Care and Use Committee (IACUC) and were in accordance with the Association for Research in Vision and Ophthalmology Statement for the use of Animals in Ophthalmic and Visual Research.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2020.106076.
Appendix. Supplementary materials
References
- 1.Zhong Y., Mohan K., Liu J., Al-Attar A., Lin P., Flight R.M., Sun Q., Warmoes M.O., Deshpande R.R., Liu H., Jung K.S., Mitov M.I., Lin N., Butterfield D.A., Lu S., Liu J., Moseley H.N.B., Fan T.W.M., Kleinman M.E., Wang Q.J. Loss of CLN3, the gene mutated in juvenile neuronal ceroid lipofuscinosis, leads to metabolic impairment and autophagy induction in retinal pigment epithelium. Biochim. Biophys. Acta. Mol. Basis Dis. 2020 doi: 10.1016/j.bbadis.2020.165883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cotman S.L., Vrbanac V., Lebel L.A., Lee R.L., Johnson K.A., Donahue L.R., Teed A.M., Antonellis K., Bronson R.T., Lerner T.J., MacDonald M.E. Cln3(Deltaex7/8) knock-in mice with the common JNCL mutation exhibit progressive neurologic disease that begins before birth. Hum. Mol. Genet. 2002;11:2709–2721. doi: 10.1093/hmg/11.22.2709. [DOI] [PubMed] [Google Scholar]
- 3.Chang B., Hawes N.L., Hurd R.E., Davisson M.T., Nusinowitz S., Heckenlively J.R. Retinal degeneration mutants in the mouse. Vision Res. 2002;42:517–525. doi: 10.1016/s0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- 4.Mattapallil M.J., Wawrousek E.F., Chan C.C., Zhao H., Roychoudhury J., Ferguson T.A., Caspi R.R. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest. Ophthalmol. Vis. Sci. 2012;53:2921–2927. doi: 10.1167/iovs.12-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleine Holthaus S.M., Aristorena M., Maswood R., Semenyuk O., Hoke J., Hare A., Smith A.J., Mole S.E., Ali R.R. Gene therapy targeting the inner retina rescues the retinal phenotype in a mouse model of CLN3 Batten disease. Hum. Gene. Ther. 2020 doi: 10.1089/hum.2020.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudolf M., Vogt S.D., Curcio C.A., Huisingh C., McGwin G., Jr., Wagner A., Grisanti S., Read R.W. Histologic basis of variations in retinal pigment epithelium autofluorescence in eyes with geographic atrophy. Ophthalmology. 2013;120:821–828. doi: 10.1016/j.ophtha.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall N.A., Lake B.D., Dewji N.N., Patrick A.D. Lysosomal storage of subunit c of mitochondrial ATP synthase in Batten's disease (ceroid-lipofuscinosis) Biochem. J. 1991;275(Pt 1):269–272. doi: 10.1042/bj2750269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer D.N., Fearnley I.M., Walker J.E., Hall N.A., Lake B.D., Wolfe L.S., Haltia M., Martinus R.D., Jolly R.D. Mitochondrial ATP synthase subunit c storage in the ceroid-lipofuscinoses (Batten disease) Am. J. Med. Genet. 1992;42:561–567. doi: 10.1002/ajmg.1320420428. [DOI] [PubMed] [Google Scholar]
- 9.Staropoli J.F., Haliw L., Biswas S., Garrett L., Holter S.M., Becker L., Skosyrski S., Da Silva-Buttkus P., Calzada-Wack J., Neff F., Rathkolb B., Rozman J., Schrewe A., Adler T., Puk O., Sun M., Favor J., Racz I., Bekeredjian R., Busch D.H., Graw J., Klingenspor M., Klopstock T., Wolf E., Wurst W., Zimmer A., Lopez E., Harati H., Hill E., Krause D.S., Guide J., Dragileva E., Gale E., Wheeler V.C., Boustany R.M., Brown D.E., Breton S., Ruether K., Gailus-Durner V., Fuchs H., de Angelis M.H., Cotman S.L. Large-scale phenotyping of an accurate genetic mouse model of JNCL identifies novel early pathology outside the central nervous system. PLoS One. 2012;7:e38310. doi: 10.1371/journal.pone.0038310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanks J.C., Johnson L.V. Specific binding of peanut lectin to a class of retinal photoreceptor cells. A species comparison. Invest. Ophthalmol. Vis. Sci. 1984;25:546–557. [PubMed] [Google Scholar]
- 11.Rubio-Fernandez M., Uribe M.L., Vicente-Tejedor J., Germain F., Susin-Lara C., Quereda C., Montoliu L., de P., Martin-Nieto J., Cruces J. Impairment of photoreceptor ribbon synapses in a novel Pomt1 conditional knockout mouse model of dystroglycanopathy. Sci. Rep. 2018;8:8543. doi: 10.1038/s41598-018-26855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao L., Li J., Fu Y., Zhang M., Wang B., Ouellette J., Shahi P.K., Pattnaik B.R., Watters J.J., Wong W.T., Guo L.W. Photoreceptor protection via blockade of BET epigenetic readers in a murine model of inherited retinal degeneration. J. Neuroinflammation. 2017;14:14. doi: 10.1186/s12974-016-0775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorval K.M., Bobechko B.P., Fujieda H., Chen S., Zack D.J., Bremner R. CHX10 targets a subset of photoreceptor genes. J. Biol. Chem. 2006;281:744–751. doi: 10.1074/jbc.M509470200. [DOI] [PubMed] [Google Scholar]
- 14.Zagozewski J.L., Zhang Q., Pinto V.I., Wigle J.T., Eisenstat D.D. The role of homeobox genes in retinal development and disease. Dev. Biol. 2014;393:195–208. doi: 10.1016/j.ydbio.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Kovacs-Valasek A., Szabadfi K., Denes V., Szalontai B., Tamas A., Kiss P., Szabo A., Setalo G., Reglodi D., Gabriel R. Accelerated retinal aging in PACAP knock-out mice. Neuroscience. 2017;348:1–10. doi: 10.1016/j.neuroscience.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Pang J.J., Yang Z., Jacoby R.A., Wu S.M. Cone synapses in mammalian retinal rod bipolar cells. J. Comp. Neurol. 2018;526:1896–1909. doi: 10.1002/cne.24456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hombrebueno J.R., Chen M., Penalva R.G., Xu H. Loss of synaptic connectivity, particularly in second order neurons is a key feature of diabetic retinal neuropathy in the Ins2Akita mouse. PLoS One. 2014;9:e97970. doi: 10.1371/journal.pone.0097970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shekhar K., Lapan S.W., Whitney I.E., Tran N.M., Macosko E.Z., Kowalczyk M., Adiconis X., Levin J.Z., Nemesh J., Goldman M., McCarroll S.A., Cepko C.L., Regev A., Sanes J.R. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016;166:1308–1323. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbato S., Marrocco E., Intartaglia D., Pizzo M., Asteriti S., Naso F., Falanga D., Bhat R.S., Meola N., Carissimo A., Karali M., Prosser H.M., Cangiano L., Surace E.M., Banfi S., Conte I. MiR-211 is essential for adult cone photoreceptor maintenance and visual function. Sci. Rep. 2017;7:17004. doi: 10.1038/s41598-017-17331-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.