Abstract
Hepatocellular carcinoma (HCC) was the third most common cause of cancer-associated mortality in China in 2015. Early detection of HCC and hepatic cirrhosis (HC) can serve a crucial role in the prevention and therapeutic intervention of these diseases. Current early detection methods rely on less sensitive imaging modalities compared with the pathological examination. In the present study, a total of 64 patients with HCC, 44 patients with HC and 298 individuals with no evidence of disease (NED) were recruited, and the ability of methylated septin 9 (mSEPT9) in diagnosing HCC and HC was investigated. The overall detection sensitivity of mSEPT9 for HCC and HC was 76.7 and 34.1%, respectively, with a 95.9% specificity (HCC vs. NED). The sensitivity of mSEPT9 for HCC was significantly higher than that of α-fetoprotein (AFP; χ2 test; 56.7%; P<0.05). The areas under the curve from the receiver operating characteristic curves of mSEPT9 for detection of HCC vs. NED, HC vs. NED and HCC vs. HC were 0.85, 0.77 and 0.66, respectively, while those of AFP for the same groups were 0.80, 0.55 and 0.77, respectively. Although both markers exhibited stage-dependent sensitivity in HCC, mSEPT9 was demonstrated to be more sensitive than AFP. The net reclassification index of mSPET9 for HCC detection was 0.212 compared with AFP, suggesting an improved diagnostic performance of mSEPT9 compared with AFP. In addition, Kaplan-Meier survival analysis revealed that mSEPT9 is able to predict the long-term survival of patients with HCC. Further analysis suggested that patients >50 years of age exhibited higher sensitivity compared with those <50 years old in mSEPT9, but not in AFP. No significant difference in sensitivity was observed between compensated and decompensated patients with HC, and in patients with HC with a history of hepatitis B or C virus infection. No difference was observed between male and female subjects in the HC and HCC groups for mSEPT9 and AFP. In conclusion, mSEPT9 may detect HCC with an overall improved sensitivity compared with AFP and may help in predicting the long-term survival of patients with HCC. The present clinical study was retrospectively registered to the Chinese Clinical Trial Registry on April 4, 2020 (http://www.chictr.org.cn/enIndex.aspx; registration no. ChiCTR2000031547).
Keywords: septin 9, liver, hepatocellular carcinoma, cirrhosis, methylation
Introduction
Hepatocellular carcinoma (HCC) is the third most common type of cancer in China (1). Early detection of HCC has always been an important factor for its prevention and therapeutic intervention. Imaging modalities such as ultrasonography can be used for early screening of potentially malignant lesions and provide guidance for further examination and treatment (2). However, it is often difficult to distinguish malignant lesions from benign changes using only an imaging method. Other in vitro diagnostic methods may be required for differential diagnosis. Hepatic cirrhosis (HC) is the most common precancerous condition that may require early intervention (3). Hepatitis B virus (HBV) infection is considered as a common cause of HC in China. Other factors for HC include hepatitis C virus (HCV) infection and alcoholic cirrhosis (2). The majority of patients with HC and HCC generally exhibit >10 years of history of either hepatitis virus infection or alcohol overuse (4,5).
The plasma-based septin 9 (SEPT9) gene methylation assay, known as the SensiColon test, was approved by the Chinese Food and Drug Administration (FDA) as the first blood-based early detection test for colorectal cancer (CRC) (4,5). Multiple case-control and prospective screening studies have demonstrated its effectiveness in the early detection and screening of CRC (4,5). The assay was designed to identify the low copy number of aberrantly methylated SEPT9 (mSEPT9) DNA against the strong background of normal genomic DNA. The assay includes a high-perfromance, cell-free DNA extraction and bisulfite conversion with a single PCR reaction to measure SEPT9 methylation in plasma samples (6,7). Although the SensiColon assay was modified from the US FDA-approved Epi proColon test, which includes a triplicate PCR assay to detect mSEPT9, studies have demonstrated that it has an equal detection sensitivity and a satisfactory specificity (8,9).
It has been suggested that the sensitivity of the mSEPT9 assay is enhanced with an increase in the severity of the cancer lesions (6,8). Numerous studies have suggested that the sensitivity of the mSEPT9 assay is associated with cancer stage (6,7). Additionally, it has been demonstrated that the detection sensitivity is influenced by age, but not by sex (10). These factors are crucial for evaluating the test performance and require further investigation. Furthermore, previous studies have indicated that the aberrant blood mSEPT9 levels can be detected in other types of cancer, including lung cancer and HCC (11–13). This further suggests that mSEPT9 detection may be used in the diagnosis of multiple types of cancer, especially in combination with other markers.
In the present study, a case-control study was performed to evaluate the performance of the mSEPT9 assay in patients with HCC and HC compared with the performance of α-fetoprotein (AFP). The performance of the combination of the two approaches was also examined. The association between mSEPT9 detection and HCC prognosis was investigated to determine whether mSEPT9 could predict the long-term survival of patients with HCC.
Materials and methods
Ethics
A detailed plan for the present study was submitted to the Ethics Committees of the First Affiliated Hospital of Xian Medical University (Xian, China) and Wuqi People's Hospital (Yanan, China) for review and was approved prior to the initiation of the clinical study. All subjects involved in the present study provided written informed consent before blood collection, and were informed of the usage of plasma and of the test results. Confirmation of approval for clinical studies was received from the institutional review board or ethics committee of the aforementioned hospitals. The present clinical study was retrospectively registered to the Chinese Clinical Trial Registry on April 4, 2020 (http://www.chictr.org.cn/enIndex.aspx; registration no. ChiCTR2000031547).
Study design, patients and blood collection
The present case-control study was designed and performed at the First Affiliated Hospital of Xian Medical University and Wuqi People's Hospital using the mSEPT9 assay (SensiColon; BioChain Institute, Inc.). The clinical status of all subjects was determined before blood collection for the mSEPT9 assay, and blood samples were obtained from all subjects who met the selection criteria between June 2016 and April 2017 in the two aforementioned hospitals. A total of 406 subjects were enrolled in the present study, including 64 patients with HCC (mean age, 58 years; age range, 33–77 years), 44 patients with HC (mean age, 56 years; age range, 33–84 years) and 298 healthy individuals with no evidence of disease (NED; Table I) (mean age, 53 years; age range, 17–88 years). The classification of all conditions was based on diagnosis from ultrasonic and computed tomography (CT) scans, and subsequent pathological examinations. Patients with HCC were divided into three subgroups according to stage (A, B or C) based on the Barcelona Clinic Liver Cancer (BCLC) staging system (14), and patients with HC were divided into two categories (compensated and decompensated) based on the assessment of patient condition, as previously described (15). The BCLC staging system was used since it takes into consideration the liver function and was capable of predicating the prognosis, and the status of mSEPT9 is closely associated with the functional change and is predictive for survival (13), while the TNM staging does not consider these factors.
Table I.
Numbers of enrolled individuals according to diagnosis group.
| Sex | Age, years | |||||||
|---|---|---|---|---|---|---|---|---|
| Diagnosis group | Total, n | Male | Female | <50 | 50-59 | 60-69 | ≥70 | |
| Total, n | 406 | 303 | 103 | 165 | 126 | 70 | 45 | |
| HCC | 64 | 50 | 14 | 16 | 19 | 19 | 10 | |
| Stage A | 23 | 17 | 6 | 6 | 6 | 8 | 3 | |
| Stage B | 23 | 19 | 4 | 5 | 7 | 7 | 4 | |
| Stage C | 18 | 14 | 4 | 5 | 6 | 4 | 3 | |
| HC | 44 | 33 | 11 | 17 | 16 | 7 | 4 | |
| Decompensated | 11 | 7 | 4 | 5 | 3 | 2 | 1 | |
| Compensated | 33 | 26 | 7 | 12 | 13 | 5 | 3 | |
| NED | 298 | 220 | 78 | 132 | 91 | 44 | 31 | |
HCC, hepatocellular carcinoma; HC, hepatic cirrhosis; NED, no evidence of disease.
The inclusion criteria were that subjects had to be adults >18 years old with complete clinicopathological information and confirmed diagnosis of HCC, HC or NED (healthy subjects) via imaging examination (including endoscopy, ultrasound, magnetic resonance imaging and CT scans) and/or subsequent pathological examination. The exclusion criteria included: Pregnant women, history of any type of cancer or history of therapy for any type of cancer. Subjects with incomplete clinicopathological or follow-up information were also excluded. The NED group was recruited from asymptomatic individuals who came to the hospitals for physical examinations, and the HC group was recruited from symptomatic individuals with known liver diseases (not malignant liver diseases) who came to the hospitals for diagnosis and therapy. All subjects underwent blood collection before examination and before subsequent biopsies or surgery were performed. None of the subjects received chemotherapy, radiotherapy or surgical intervention before blood collection. All patients were followed up to 630 days.
Sample size estimation
Sample size estimation was based on the following equation for known detection sensitivity: N=Z2x[p(1-p)]/E2. The parameters were defined as follows: Z is a statistical parameter (Z=1.96 for 95% CI), E represents the error (10% was selected for the present study) and p represents the putative positive detection rate (PDR). A P-value of 0.80 was obtained from our previous pilot study investigating the sensitivity of the mSEPT9 assay in HCC (data not published). It was estimated that 61 patients with HCC were required. Using the same method, the number of patients with HC was also estimated. When P=0.12, the number of patients with HC required was 41. Healthy subjects were recruited based on the ratio of HCC:NED (1:2), and therefore the study goal was to recruit 122 healthy subjects, with a total of 224 individuals.
Sample collection and storage
Samples were collected from outpatients and inpatients, and sample information was recorded in sample collection forms. A 10-ml peripheral blood sample was collected in 10 ml K2-EDTA anticoagulant tubes (BD Biosciences) to ensure the accuracy of the assay. Sample storage and transportation were performed following the manufacturers protocol of the SensiColon assay.
DNA extraction, quantitative PCR analysis of SEPT9 and the AFP test
DNA extraction from 10 ml plasma samples (circulating tumor DNA) and bisulfite conversion were performed according to the manufacturers protocol of the SensiColon assay [BioChain (Beijing) Science and Technology, Co., Ltd.]. The bisulfite-DNA was assayed with SensiColon kits on an ABI 7500 Fast Dx Real Time PCR device (Thermo Fischer Scientific, Inc.). Primers and conditions for qualitative PCR analysis of mSEPT9 were used according to previously described methods (6,8–10). Briefly, PCR was performed in triplicate with 15 µl template DNA per well and run for 45 cycles. PCR results for β-actin (ACTB) and mSEPT9 for each of the triplicate reactions were recorded using the instrument software. PCR was performed using a Taqman-based assay (Roche Diagnostics) with fluorophore detection. The sequence of primers, blockers and probes for SEPT9 detection used in methylation-specific PCR amplification are as follows: Forward primer, 5-CCCACCAACCATCATAT-3 and reverse primer, 5-GTAGTAGTTAGTTTAGTATTTATTTT-3; probe1, 5-GTTCGAAATGATTTTATTTAGTTGC-3; and probe2, 5-CGTTGATCGCGGGGTTC-3 (9). β-actin was used as the internal control to evaluate the plasma DNA quality and the validity of PCR amplification. The sequence of primers and probes for β-actin detection used in PCR amplification were as follows: Forward, 5-GTGATGGAGGAGGTTTAGTAAGTT-3 and reverse, 5-CCAATAAAACCTACTCCTCCCTTAA-3; and probe, 5-ACCACCACCCAACACACAATAACAAACACA-3 (9). The thermocycling conditions were as follows: Initial denaturation at 94°C for 20 min, followed by 45 cycles at 62°C for 5 sec, 55.5°C for 35 sec and 93°C for 30 sec; and cooling at 40°C for 5 sec. The validity of each sample batch was determined on the basis of mSEPT9 and ACTB threshold count (Cq) values for the positive and negative controls (16). DNA from treated Jurkat cells was used as the positive control and from treated HeLa cells was used as the negative control. ACTB was used as an internal reference to assess the integrity of each sample. The AFP assay used was a commercial test adopted by the participating hospital, and its cut-off was set to 20 µg/l according to the manufacturers protocol [Roche Diagnostics (Shanghai) Co., Ltd.].
Data analysis and interpretation
The data from the PCR reactions of the SensiColon assay were analyzed using the 1/1 algorithm, which means that a sample was considered to be positive if the only PCR was positive and was considered to be negative if it was negative (8,9). The detailed methods for data analysis and interpretation have been previously described (6,8–10). For each sample, a relative methylation value was determined using the 2−ΔΔCq method adapted for DNA methylation analyses as previously described (16). In brief, ΔΔCq values were calculated as follows: ΔΔCqSample=ΔCqSample-ΔCqCalibrator, where ΔCqSample=CqACTB of sample-CqSEPT9 of sample and ΔCqCalibrator=CqACTB of calibrator-CqSEPT9 of calibrator. The cut-off of the SensiColon assay in HCC detection was re-defined using receiver operating characteristic (ROC) curves and the ΔΔCq values of the control and the HCC group, and was determined by the best balance between sensitivity and specificity. The cut-off was set to a Cq value of 41.1 (ΔΔCq=−4.0) based on the aforementioned considerations.
Statistical analysis
Analyses including unpaired Students t-test for comparison of two groups with normal distribution (experiments were repeated four times to ensure the repeatability and stability of the test), one-way ANOVA followed by a post hoc test (Bonferronis correction) for comparison of >3 groups with normal distribution and χ2 test for comparison of rate or percentage between groups. ROC curves and Kaplan-Meier survival curves were performed using GraphPad Prism 5.0 (GraphPad Software, Inc.). Log-rank (Mantel-Cox) test was used to compare two survival curves. Bonferroni correction was performed for the P-values from χ2 tests when >2 groups were compared using multiple 2×2 χ2 tests. P<0.05, P<0.0167, P<0.0125 or P<0.01 were considered to indicate statistically significant differences when comparing 2, 3, 4 or 5 groups, respectively. The positive predictive value (PPV)=number of true positives/total number of positives, and the negative predictive value (NPV)=number of true negatives/total number of negatives. The positive likelihood ratio (+LR)=sensitivity/(1-specificity), and the negative likelihood ratio (−LR)=(1-sensitivity)/specificity. The net reclassification index (NRI)=(sensitivitySEPT9 + specificitySEPT9)-(sensitivityAFP + specificityAFP). Data are presented in scatter plots or histograms with mean values and 95% CI where appropriate, and in ROC curves or Kaplan-Meier survival curves where appropriate. The number of subjects for each statistical group was shown in Table I.
Results
mSEPT9 exhibits an improved overall performance in HCC detection compared with AFP
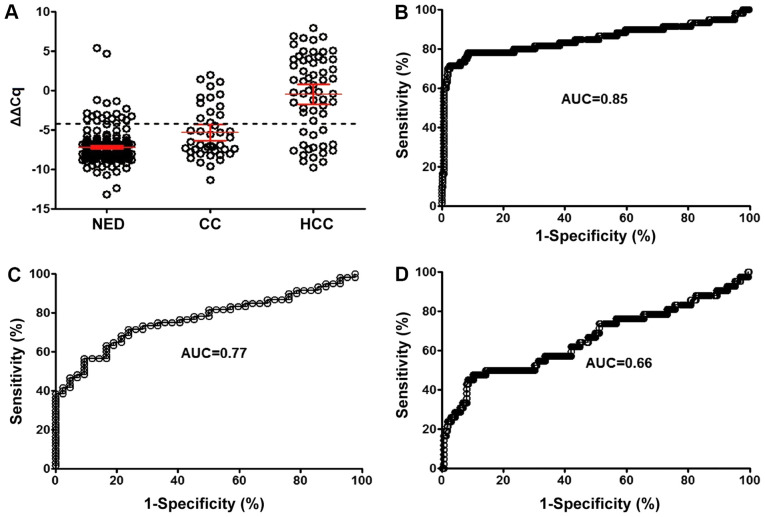
In order to investigate the diagnostic sensitivity of mSEPT9 in HCC and HC, the threshold value of mSEPT9 detection was defined by examining plasma samples from individuals with HCC, HC and NED. Combined analysis of ΔΔCq value distribution and ROC curve (Fig. 1A and B) suggested that the optimal sensitivity (76.7%) and specificity (95.6%) of the mSEPT9 assay for HCC were obtained at a Cq threshold of 41.1 (ΔΔCq=−4.0), with an area under the curve (AUC) of 0.85 (95% CI, 0.78-0.92). Under the same threshold, the detection sensitivity for HC was 34.1% with an AUC of 0.77 (95% CI, 0.68-0.86; Fig. 1C), while the AUC for discrimination between HCC and HC was 0.66 (95% CI, 0.56-0.76; Fig. 1D). The PPV for the mSEPT9 assay was 63.0%, and the NPV was 86.8%. The +LR was 18.7 and the -LR was 0.243, suggesting a high true positive probability when a test was positive, and a high true negative probability when a test was negative (data not shown). The NRI of mSPET9 for HCC was 0.212 compared with AFP, suggesting an improved diagnostic performance of mSEPT9 compared with AFP.
Figure 1.
Scatter plot and ROC curves for HCC and HC detection using the mSEPT9 assay. (A) Scatter plot showing the mSEPT9 ΔΔCq values from the blood of individuals with NED, HC and HCC. ROC curves for mSEPT9 sensitivity in detecting (B) HCC vs. NED, (C) HC vs. NED and (D) HCC vs. HC. The threshold (cutoff value) for ΔΔCq (dotted line) was set at −4.0. Bars represent the mean ± 95% CI. mSEPT9, methylated septin 9; ROC, receiver operating characteristic; HCC, hepatocellular carcinoma; HC, hepatic cirrhosis; NED, no evidence of disease; AUC, area under the curve.
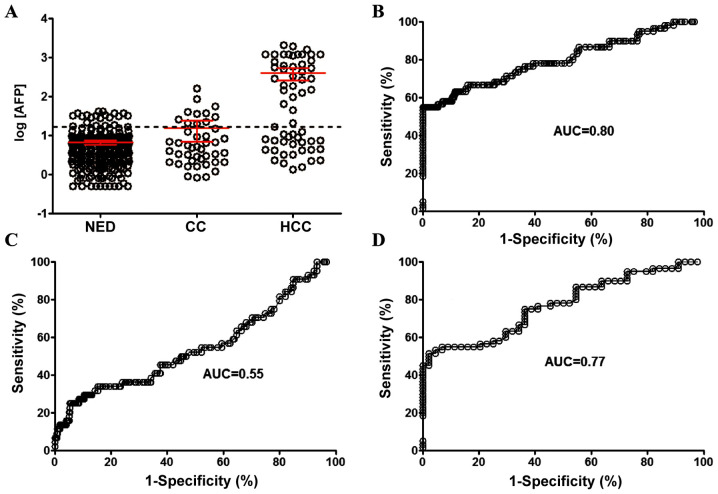
The detection sensitivity of AFP in the same groups of individuals with HCC, HC and NED was also examined. When the threshold for discriminating positive or negative detection was set at 20 ng/ml (Fig. 2A), the detection sensitivity for HCC was 56.7% with an AUC of 0.80 (95% CI, 0.73-0.87; Fig. 2B), and the sensitivity of HC detection was 25.0% with an AUC of 0.55 (95% CI, 0.45-0.65; Fig. 2C). The AUC for discrimination between HCC and HC was 0.77 (95% CI, 0.68-0.86; Fig. 2D).
Figure 2.
Scatter plot and ROC curves for HCC and HC detection using AFP. (A) Scatter plot showing the serum AFP concentration of individuals with NED, HC and HCC. ROC curves for AFP sensitivity in detecting (B) HCC vs. NED, (C) HC vs. NED and (D) HCC vs. HC. The threshold (cutoff value) for AFP (dotted line) was set at 20 µg/l (log value at 1.30). Bars represent the mean ± 95% CI. AFP, α-fetoprotein; ROC, receiver operating characteristic; HCC, hepatocellular carcinoma; HC, hepatic cirrhosis; NED, no evidence of disease; AUC, area under the curve.
Combination of mSEPT9 and AFP enhances HCC detection performance
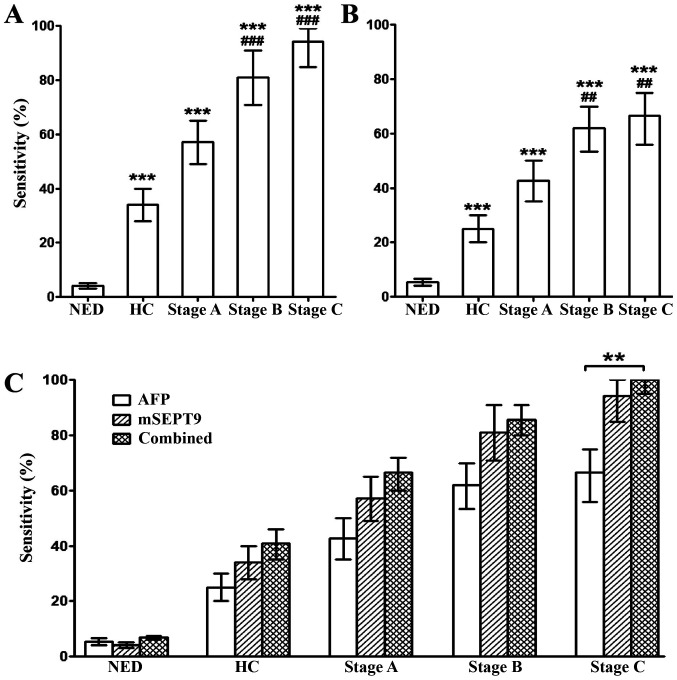
Furthermore, the stage-dependent sensitivity for HCC in mSEPT9 and AFP detection was examined. The sensitivity of mSEPT9 for NED, HC and Stage A, B and C HCC was 4.1, 34.1, 57.1, 81.0 and 94.4%, respectively, at 95.9% specificity (Fig. 3A), while the sensitivity of AFP for the aforementioned groups was 5.4, 25.0, 42.8, 61.9 and 66.7%, respectively, at 94.6% specificity (Fig. 3B). Combined detection of mSPET9 and AFP significantly enhanced the detection sensitivity compared with AFP alone in stage C HCC (P<0.01), while a trend of increased sensitivity was also observed in HC and stage A and B HCC, although the differences were not statistically significant (Fig. 3C). In addition, the combined sensitivity was not statistically different compared with the sensitivity of mSEPT9 alone in the diagnosis of the aforementioned groups (Fig. 3C).
Figure 3.
Sensitivity of mSEPT9 alone, AFP alone and both combined for the detection of NED, HC and stage A, B and C HCC. Sensitivity of (A) mSEPT9, (B) AFP and (C) both combined. Bonferroni correction was performed for the P-values from the χ2 tests when >2 groups were compared. **P<0.01; ***P<0.001 vs. NED; and ##P<0.01; ###P<0.001 vs. HC. mSEPT9, methylated septin 9; AFP, α-fetoprotein; HCC, hepatocellular carcinoma; HC, hepatic cirrhosis; NED, no evidence of disease.
mSEPT9 detection predicst long-term survival in patients with HCC
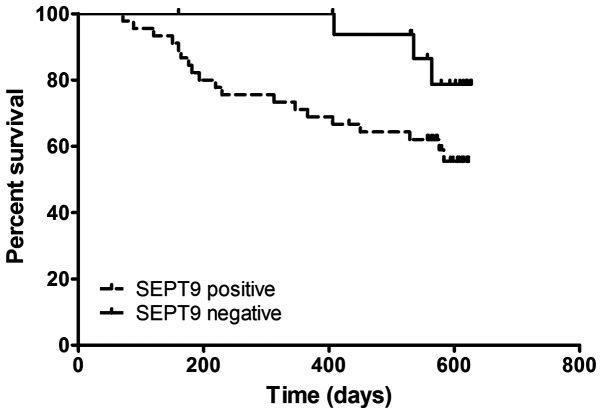
To investigate the potential role of mSEPT9 in the prognosis of patients with HCC, these patients were followed for ~630 days (21 months) and a Kaplan-Meier survival analysis was performed. As indicated in Fig. 4, the Kaplan-Meier curves were plotted according to mSEPT9 positive or negative detection. Patients with positive mSEPT9 detection exhibited a significantly poorer survival rate than patients with negative mSEPT9 (P=0.039; hazard ratio, 2.343; 95% CI, 0.957-5.736). The present findings suggested that mSEPT9 may predict the long-term survival of patients with HCC, regardless of the disease stage or therapy.
Figure 4.
Kaplan-Meier curves for the survival of patients with hepatocellular carcinoma based on mSEPT9 positive or negative detection. Patients were considered as mSEPT9 positive or negative based on a Cq threshold of 41.1. mSEPT9, methylated septin 9.
Diagnostic accuracy of mSEPT9 is influenced by age, but not by sex or type of hepatitis virus infection
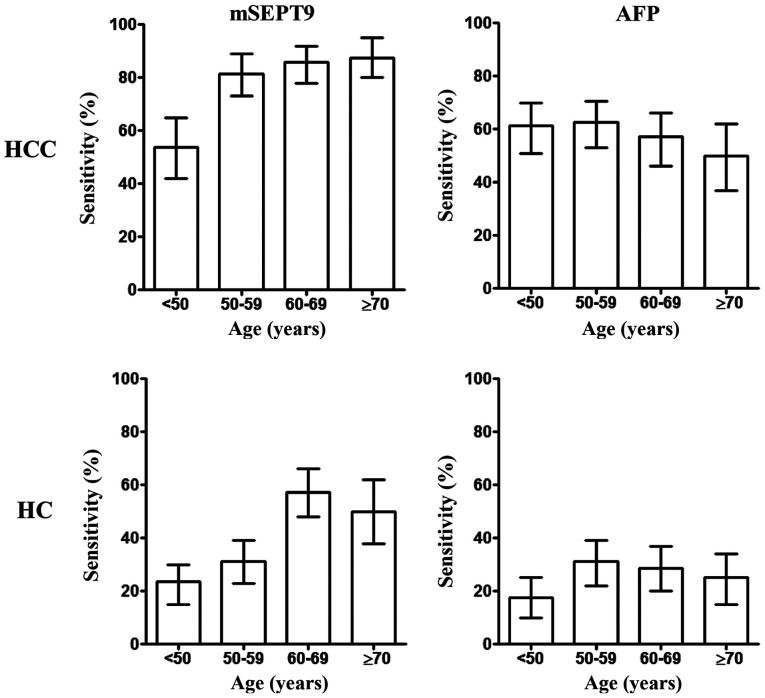
In order to investigate other factors influencing the diagnostic accuracy of mSEPT9 in HCC, information about patient age, sex and hepatitis infection status was collected and the association between these factors and mSEPT9 detection performance was analyzed (Fig. 5), which suggested a higher sensitivity in patients >50 years old (including groups: 50–59, 60–69 and ≥70 years; 84.2%) compared with those <50 years (53.8%) (P=0.026; data not shown). Similarly, a higher sensitivity in HC detection was observed in all patients >60 years old (including groups: 60–69 and ≥70 years; 27.3%) (P=0.098; data not shown) compared with those <60 years of age (including groups: <50 and 50–59 years; 54.5%). By contrast, no significant differences in sensitivity were detected across the various age groups with AFP detection.
Figure 5.
Detection sensitivity for HCC and HC using mSEPT9 or AFP levels according to age. mSEPT9, methylated septin 9; AFP, α-fetoprotein; HCC, hepatocellular carcinoma; HC, hepatic cirrhosis.
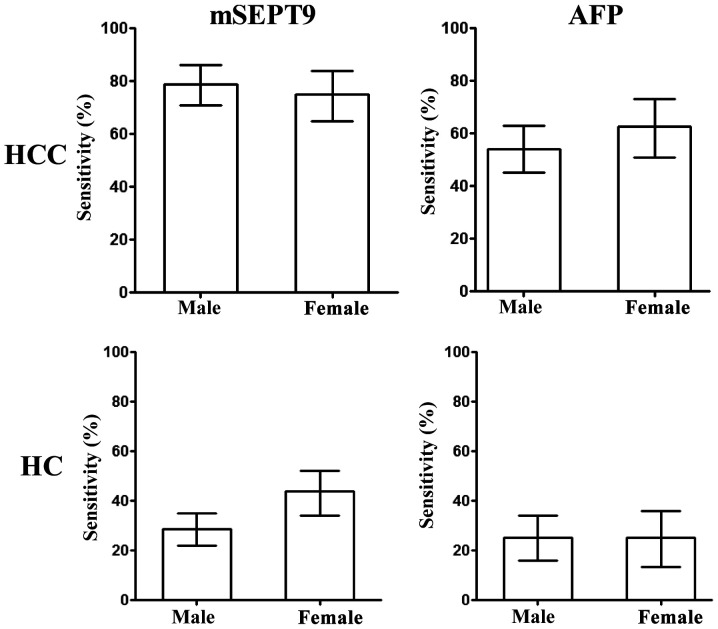
Furthermore, the influence of the sex of the patients on HCC and HC diagnosis using mSEPT9 and AFP detection was analyzed. As revealed in Fig. 6, no significant differences were observed between males and females in HCC or HC detection with either mSEPT9 or AFP, indicating a non-discriminative detection for both sexes.
Figure 6.
Detection sensitivity for HCC and HC using mSEPT9 or AFP levels according to sex. mSEPT9, methylated septin 9; AFP, α-fetoprotein; HCC, hepatocellular carcinoma; HC, hepatic cirrhosis.
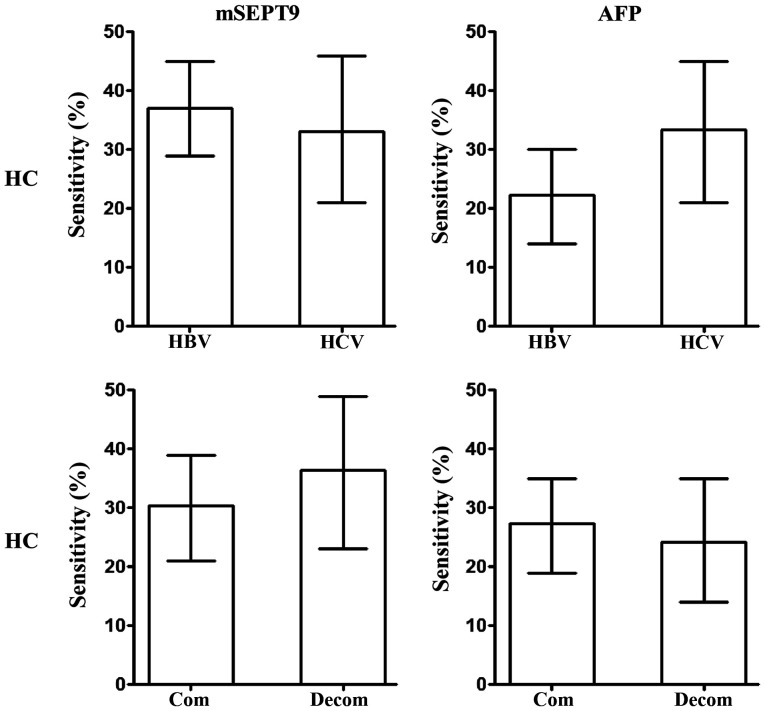
The majority of patients with HC have a history of hepatitis virus infection, commonly HBV infection followed by HCV infection (4,5). Therefore, the potential difference between HBV- and HCV-infected patients with HC was analyzed. As demonstrated in Fig. 7, no difference in the sensitivity of HC diagnosis between HBV- and HCV-infected patients was observed using mSEPT9 or AFP detection. Additionally, since both compensated and decompensated patients with HC are often observed clinically, the potential difference in the detection sensitivity between these two groups was analyzed. The present results suggested no significant differences between the sensitivity of either mSEPT9 or AFP detection in patients with compensated and decompensated HC (Fig. 7).
Figure 7.
Effect of HBV or HCV infection on HC and compensation status on HC detection sensitivity using mSEPT9 and AFP levels. HBV/HCV, hepatitis B/C virus; com, compensated; decom, decompensated; mSEPT9, methylated septin 9; AFP, α-fetoprotein; HC, hepatic cirrhosis.
Discussion
The mSEPT9 assay has been used in the screening and early detection of CRC for >5 years. The assay has been approved by the US FDA as a screening test for the average-risk population >50 years of age and by the Chinese FDA as an early detection test for high-risk populations (9,17) as a proven specific test for CRC. Additionally, it can effectively detect colorectal precancerous conditions, such as adenoma (18,19). Furthermore, aberrant mSEPT9 levels have been detected in other types of cancer, such as lung cancer and HCC (9–11), where they may serve as markers.
The performance of mSEPT9 and AFP assays in HC and HCC detection has been investigated in a previous study conducted in a European population using the Epi proColon 2.0 CE assay (13). The Epi proColon 2.0 CE test performs three parallel PCR reactions to detect the aberrant methylation levels of the SEPT9 gene, which requires an appropriate algorithm for result interpretation. The findings of the present study suggest that the mSEPT9 assay may be used to detect HCC and HC with high sensitivity in a Chinese population. In contrast to a previous study, a modified version of the mSEPT9 assay approved by the Chinese FDA for CRC early detection (9) was used to validate its applicability for the diagnosis of HCC and HC. The modified mSEPT9 assay allowed the detection of aberrant mSEPT9 with a single PCR reaction, therefore facilitating its use in routine laboratory testing. The overall sensitivity for HCC diagnosis (76.7%) was similar to that for CRC diagnosis (9), and it was the same as that reported for HCC in a European population (13), indicating that mSEPT9 may also detect HCC with a high sensitivity in a Chinese population. Since the assay was initially designed and validated in CRC, the present findings suggested that CRC and HCC may have similar mSEPT9 patterns, although this requires further study. Furthermore, the present results indicated that mSEPT9 was not only specific to CRC but can also be detected in other cancer types, such as HCC. Additionally, a cancer stage dependent sensitivity of mSEPT9 was observed, starting from benign HC to malignant HCC stages A-C. This further indicated that mSEPT9 levels may reflect the severity of the progression of hepatic lesions, in agreement with previous studies on CRC where a stage-dependent sensitivity was observed (6,10). Therefore, mSEPT9 levels may also be used in HC or HCC therapeutic response assessment or monitoring, as this application has been validated in surgery and late-stage CRC therapies (20).
Notably, the present study revealed that the mSEPT9 assay had a higher sensitivity in patients >50 years of age than in younger patients. This may reflect an increase in mSEPT9 levels with increasing age in patients with HCC, which was also observed in patients with CRC (4,5,10), suggesting a common trend associated with increasing age. In patients with CRC, a significant difference in positive detection rate was observed between individuals >60 and <60 years of age, which may be attributed to the late-onset age of CRC (4,5,10) compared with that of HCC. Changes in gene methylation status with age can occur in cancer as well as in normal tissues. It has been demonstrated that low-level de novo methylation of CpG islands occur in normal tissues and that their frequency increases with age (21–23). Abnormal methylation levels observed in tumor cells can also occur in normal cells (24). Overall, aberrant methylation patterns detected in cancer may be present in normal aging cells, which may result in malignant transformation due to the accumulation of methylation aberrances with increasing age.
The present study suggested that the sex of the patients did not influence the sensitivity of the mSEPT9 assay, indicating that both sexes may share a similar mSEPT9 gene profile and that the mechanism of HCC carcinogenesis may be similar between them. This would further increase the applicability of the mSEPT9 assay for HCC detection.
The present findings suggested that the diagnostic performance of mSEPT9 was improved compared with that of AFP in the detection of HCC (sensitivity, 76.7 vs. 56.7%; AUC, 0.85 vs. 0.80) and HC (sensitivity, 34.1 vs. 25.0%; AUC, 0.77 vs. 0.55), with no marked difference in their specificity (95.9 vs. 94.6%). Additionally, a higher sensitivity of mSEPT9 compared with AFP was observed in diagnosing all HCC stages. By contrast, the performance of mSEPT9 in discriminating HCC from HC was not higher compared with that of AFP (AUC, 0.66 vs. 0.77). The present observations suggested that mSEPT9 may be more suitable than AFP as a marker for HCC detection. However, it was less effective in the differential diagnosis between HCC and HC, possibly due to the higher detection rate of HC by mSEPT9 increasing the difficulty in discriminating it from HCC. A previous study has reported a similar behavior of mSEPT9 when one marker is sensitive to a certain type of cancer and its precancerous conditions (18). Furthermore, the present findings demonstrated that abnormal mSEPT9 levels may also occur in HC, suggesting that aberrant methylation may exist when lesions are at the benign stage. The transition from benign HC to malignant HCC may be due to quantitative accumulation of aberrancies that ultimately lead to malignant transformation.
The combination of mSEPT9 and carcinoembryonic antigen detection enhances the diagnostic performance of these tests in CRC (9). However, in the present study, the combination of mSEPT9 and AFP detection did not significantly alter the performance of these tests in HCC and HC compared with mSEPT9 detection alone. This may be due to the fact that the proportion of overlapping detection between mSEPT9 and AFP was substantial, which may result in insignificant complementation. The present study suggested that detection of mSEPT9 alone may be enough for HCC diagnosis. By contrast, a trend of increased sensitivity was observed when the combination was compared with the detection of AFP alone, suggesting that the combination may potentially improve the detection performance of AFP compared with using this protein marker alone. Furthermore, mSEPT9 detection may be used in combination with AFP detection in the differential diagnosis between HCC and HC, as the latter appeared more potent in discriminating HCC from HC. In clinical practice, mSEPT9 and/or AFP detection may be used in combination with ultrasonic examination to support the differential diagnosis. Ultrasonic examination can discriminate the majority of HC from HCC cases (25), and mSEPT9 and/or AFP may be used for the remaining uncertain cases to maximize the definite diagnosis and reduce misdiagnoses.
An important finding of the present study was that the survival of patients with HCC may be predicted using mSEPT9 detection. Similar observations were reported for mSEPT9 in patients with CRC and for methylated short stature homeobox 2 in patients with lung cancer, independently of disease stage and therapeutic interventions (20,26). Additionally, previous studies have suggested that mSEPT9 may serve as an independent risk factor for CRC (208) and HCC (13). Therefore, mSEPT9 may be used to predict the long-term risk of patients with CRC and HCC independently. AFP has been proposed as a predictive marker for HCC in multiple studies (27,28). However, due to the lower sensitivity of AFP detection in patients with HCC, only patients with a positive AFP detection before therapy can be assessed, while patients exhibiting negative AFP detection are often hard to assess (29). By contrast, the positive detection rate of mSEPT9 was higher than that of AFP (Fig. 3C), and may therefore be more applicable in the assessment of prognosis or in predicting survival. This would be another advantage of mSEPT9 over AFP detection, especially for in vitro HCC diagnosis, in addition to its higher sensitivity. Since the promoter region of SEPT9 was reported to be highly methylated in HCC in a previous study (13) and in the present study, it is expected that the expression levels of SEPT9 would be decreased, as it was demonstrated that hypermethylation of the promoter region of SEPT9 decreased SEPT9 expression in CRC (12,30,31).
There are a few of limitations to the present study. Firstly, this was a case-control study, and thus whether mSEPT9 will exhibit a similar performance in a screening setting requires further investigation. Moreover, it was difficult to discriminate HCC from HC using mSEPT9, and it appeared that mSEPT9 alone may not be sufficient for the differential diagnosis. High sensitivity of mSEPT9 in both CRC and HCC means it is difficult to distinguish the two cancer types in screening, and other methods should be combined to overcome this issue. Future studies should focus on studying the applicability of mSEPT9 as a predictive marker for long-term prognosis in patients with HC, as ~34.1% of patients with HC in the present study were mSEPT9 positive. Potential differences in the prognosis or therapeutic response based on mSEPT9 dichotomization may help in distinguishing different patients with HC and in developing personalized therapeutic strategies. This would greatly benefit patients with HC and may prevent its progression to HCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the project ‘The key molecular events in the carcinogenesis and development of gastrointestinal cancer and the clinical significance’ funded by the Natural Science Foundation of China for innovative research group (grant no. 81421003).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors contributions
YY and NH designed the study. NH, GF, CZ, FW and TZ recruited the subjects and collected the samples. FW and TZ arranged the test of samples and liaised with the laboratory. NH, GF and CZ, FW and TZ analyzed the data. NH, GF, CZ, FW and TZ made the figures and tables, and wrote the manuscript. YY proofread the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
A detailed plan of the present study was submitted to the Ethics Committee of the First Affiliated Hospital of Xian Medical University (Xian, China) and Wuqi People's Hospital (Yanan, China) for review, and was approved before the initiation of the clinical study. All subjects involved in the study provided written informed consent before blood collection, and were informed of the usage of plasma and the test results. Confirmation of approval for clinical studies was received from the institutional review board or ethics committee of the aforementioned hospitals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:2705–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Hung CH, Lu SN, Wang JH, Lee CM, Chen TM, Tung HD, Chen CH, Huang WS, Changchien CS. Correlation between ultrasonographic and pathologic diagnoses of hepatitis B and C virus-related cirrhosis. J Gastroenterol. 2003;38:153–157. doi: 10.1007/s005350300025. [DOI] [PubMed] [Google Scholar]
- 3.Karhunen PJ, Penttilä A. Preneoplastic lesions of human liver. Hepatogastroenterology. 1987;34:10–15. [PubMed] [Google Scholar]
- 4.Alberti A, Chemello L, Benvegnù L. Natural history of hepatitis C. J Hepatol. 1999;1(Suppl 31):S17–S24. doi: 10.1016/S0168-8278(99)80369-9. [DOI] [PubMed] [Google Scholar]
- 5.Lyu X, Liu K, Chen Y, Wang Z, Yao J, Cai G, Jiang Z, Wang Z, Jiang J, Gu H. Analysis of risk factors associated with the development of hepatocellular carcinoma in chronic HBV-infected Chinese: A meta-analysis. Int J Environ Res Public Health. 2016;13:604. doi: 10.3390/ijerph13060604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song L, Jia J, Peng X, Xiao W, Li Y. The performance of the SEPT9 gene methylation assay and a comparison with other CRC screening tests: A meta-analysis. Sci Rep. 2017;7:3032. doi: 10.1038/s41598-017-03321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song L, Li Y. Progress on the clinical application of the SEPT9 gene methylation assay in the past 5 years. Biomark Med. 2017;11:415–418. doi: 10.2217/bmm-2017-0091. [DOI] [PubMed] [Google Scholar]
- 8.Song L, Li Y, Jia J, Zhou G, Wang J, Kang Q, Jin P, Sheng J, Cai G, Cai S, Han X. Algorithm optimization in methylation detection with multiple RT-qPCR. PLoS One. 2016;11:e0163333. doi: 10.1371/journal.pone.0163333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D, Zhou G, Jin P, Zhu J, Li S, Wu Q, Wang G, Sheng J, Wang J, Song L, et al. Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn. 2016;18:535–545. doi: 10.1016/j.jmoldx.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Song L, Jia J, Yu H, Peng X, Xiao W, Gong Y, Zhou G, Han X, Li Y. The performance of the mSEPT9 assay is influenced by algorithm, cancer stage and age, but not sex and cancer location. J Cancer Res Clin Oncol. 2017;143:1093–1101. doi: 10.1007/s00432-017-2363-0. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich D, Jung M, Puetzer S, Leisse A, Holmes EE, Meller S, Uhl B, Schatz P, Ivascu C, Kristiansen G. Diagnostic and prognostic value of SHOX2 and SEPT9 DNA methylation and cytology in benign, paramalignant and malignant pleural effusions. PLoS One. 2013;8:e84225. doi: 10.1371/journal.pone.0084225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powrózek T, Krawczyk P, Kucharczyk T, Milanowski J. Septin 9 promoter region methylation in free circulating DNA-potential role in noninvasive diagnosis of lung cancer: Preliminary report. Med Oncol. 2014;31:917. doi: 10.1007/s12032-014-0917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oussalah A, Rischer S, Bensenane M, Conroy G, Filhine-Tresarrieu P, Debard R, Forest-Tramoy D, Josse T, Reinicke D, Garcia M, et al. Plasma mSEPT9: A novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine. 2018;30:138–147. doi: 10.1016/j.ebiom.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruix J, Sherman M, Practice Guidelines Committee American Association for the Study of Liver Diseases: Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–1236. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 15.DAmico G, Morabito A, DAmico M, Pasta L, Malizia G, Rebora P, Valsecchi MG. New concepts on the clinical course and stratification of compensated and decompensated cirrhosis. Hepatol Int. 2018;12(Suppl 1):S34–S43. doi: 10.1007/s12072-017-9808-z. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rösch T, Osborn N, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–325. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song L, Peng X, Li Y, Xiao W, Jia J, Dong C, Gong Y, Zhou G, Han X. The SEPT9 gene methylation assay is capable of detecting colorectal adenoma in opportunistic screening. Epigenomics. 2017;9:599–610. doi: 10.2217/epi-2016-0146. [DOI] [PubMed] [Google Scholar]
- 19.Song L, Wang J, Wang H, Chen Y, Jia J, Guo S, Liu H, Peng X, Xiao W, Gong Y, et al. The quantitative profiling of blood mSEPT9 determines the detection performance on colorectal tumors. Epigenomics. 2018;10:1569–1583. doi: 10.2217/epi-2017-0154. [DOI] [PubMed] [Google Scholar]
- 20.Song L, Guo S, Wang J, Peng X, Jia J, Gong Y, Yang B, Xiao W, Dong C, Liu H, Li Y. The blood mSEPT9 is capable of assessing the surgical therapeutic effect and the prognosis of colorectal cancer. Biomark Med. 2018;12:961–973. doi: 10.2217/bmm-2018-0012. [DOI] [PubMed] [Google Scholar]
- 21.Kulis M, Merkel A, Heath S, Queirós AC, Schuyler RP, Castellano G, Beekman R, Raineri E, Esteve A, Clot G, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Issa JP. Aging and epigenetic drift: A vicious cycle. J Clin Invest. 2014;124:24–29. doi: 10.1172/JCI69735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JP. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagato Y, Kondo F, Kondo Y, Ebara M, Ohto M. Histological and morphometrical indicators for a biopsy diagnosis of well-differentiated hepatocellular carcinoma. Hepatology. 1991;14:473–478. doi: 10.1002/hep.1840140311. [DOI] [PubMed] [Google Scholar]
- 26.Peng X, Liu X, Xu L, Li Y, Wang H, Song L, Xiao W. The mSHOX2 is capable of assessing the therapeutic effect and predicting the prognosis of stage IV lung cancer. J Thorac Dis. 2019;11:2458–2469. doi: 10.21037/jtd.2019.05.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai DS, Zhang C, Chen P, Jin SJ, Jiang GQ. The prognostic correlation of AFP level at diagnosis with pathological grade, progression, and survival of patients with hepatocellular carcinoma. Sci Rep. 2017;7:12870. doi: 10.1038/s41598-017-12834-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan MY, She WH, Dai WC, Tsang SHY, Chok KSH, Chan ACY, Fung J, Lo CM, Cheung TT. Prognostic value of preoperative alpha-fetoprotein (AFP) level in patients receiving curative hepatectomy-an analysis of 1,182 patients in Hong Kong. Transl Gastroenterol Hepatol. 2019;4:52. doi: 10.21037/tgh.2019.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aoyagi Y, Oguro M, Yanagi M, Mita Y, Suda T, Suzuki Y, Hata K, Ichii K, Asakura H. Clinical significance of simultaneous determinations of alpha-fetoprotein and des-gamma-carboxy prothrombin in monitoring recurrence in patients with hepatocellular carcinoma. Cancer. 1996;77:1781–1786. doi: 10.1002/(SICI)1097-0142(19960501)77:9<1781::AID-CNCR4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 30.Potter NT, Hurban P, White MN, Whitlock KD, Lofton-Day CE, Tetzner R, Koenig T, Quigley NB, Weiss G. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60:1183–1191. doi: 10.1373/clinchem.2013.221044. [DOI] [PubMed] [Google Scholar]
- 31.Ravegnini G, Zolezzi Moraga JM, Maffei F, Musti M, Zenesini C, Simeon V, Sammarini G, Festi D, Hrelia P, Angelini S. Analysis of SEPT9 promoter methylation status, micronuclei frequency, and folate-related gene polymorphisms: The potential for a novel blood-based colorectal cancer biomarker. Int J Mol Sci. 2015;16:28486–28497. doi: 10.3390/ijms161226113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.