Abstract
Long non-coding RNAs (lncRNAs) represent potential biomarkers for the diagnosis and treatment of various diseases; however, the role of circulating acute ischemic stroke (AIS)-related lncRNAs remains relatively unknown. The present study aimed to screen crucial lncRNAs for AIS based on the competing endogenous RNA (ceRNA) hypothesis. The expression profile datasets for one mRNA, accession no. GSE16561, and four microRNAs (miRNAs), accession nos. GSE95204, GSE86291, GSE55937 and GSE110993, were downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs), lncRNAs (DELs), and miRNAs (DEMs) were identified, and ClusterProfiler was used to interpret the function of the DEGs. Based on the protein-protein interaction (PPI) network and module analyses, hub DEGs were identified. A ceRNA network was established based on miRNA-mRNA or miRNA-lncRNA interaction pairs. In total, 2,041 DEGs and 5 DELs were identified between the AIS and controls samples in GSE16561, and 10 DEMs between at least two of the four miRNA expression profiles. A PPI network was constructed with 1,235 DEGs, among which 20 genes were suggested to be hub genes. The hub genes paxillin (PXN), FYN-proto-oncogene, Src family tyrosine kinase (FYN), ras homolog family member A (RHOA), STAT1, and growth factor receptor-bound protein 2 (GRB2), were amongst the most significantly enriched modules extracted from the PPI network. Functional analysis revealed that these hub genes were associated with inflammation-related signaling pathways. An AIS-related ceRNA network was constructed, in which 4 DELs were predicted to function as ceRNAs for 9 DEMs, to regulate the five identified hub genes; that is, minichromosome maintenance complex component 3 associated protein-antisense RNA 1 (MCM3AP-AS1)/long intergenic non-protein coding RNA 1089 (LINC01089)/hsa-miRNA (miR)-125a/FYN, inositol-tetrakisphosphate 1-kinase-antisense RNA 1 (ITPK1-AS1)/hsa-let-7i/RHOA/GRB2/STAT1, and human leukocyte antigen complex group 27 (HCG27)/hsa-miR-19a/PXN interaction axes. In conclusion, MCM3AP-AS1, LINC01089, ITPK1-AS1, and HCG27 may represent new biomarkers and underlying targets for the treatment of AIS.
Keywords: acute ischemic stroke, competing endogenous RNAs, long non-coding RNAs, microRNAs, inflammation
Introduction
Stroke, of which ~85% are ischemic, is a common cerebrovascular disease that poses a major challenge to human health worldwide (1,2). It is estimated that 16.9 million individuals suffer a stroke every year, and ~6 million of these culminate in death, whereas three out of four survivors suffer from permanent disabilities, including motor and cognitive impairments (1,2). The occurrence of these unfavorable outcomes may be attributed to the delay in the diagnosis, and the lack of effective treatments available. Thus, in addition to conventional clinical examinations, imaging evaluation and endovascular therapy (3,4), there is an urgent need to explore novel targets for the diagnosis and treatment of acute ischemic stroke (AIS).
Long non-coding RNAs (lncRNAs), are non-coding RNAs of >200 nucleotides in length that serve important roles in various diseases, such as coronary artery disease, diabetes and cancer (5), and thus, they may represent potential biomarkers for the diagnosis and treatment of AIS. This hypothesis has been validated by numerous previous studies; for example, Zhu et al (6) identified that the expression levels of the lncRNA, myocardial infarction-associated transcript (MIAT), were significantly upregulated in blood samples compared with control samples, in addition to demonstrating that MIAT may serve as a potential diagnostic and prognostic indicator for patients with AIS [area under the curve (AUC), 0.842; sensitivity, 74.1%; specificity, 80.4%] and prognostic (AUC, 0.791; sensitivity, 79.2%; specificity, 72.9%). Feng et al (7) reported that plasma expression levels of the lncRNA, antisense non-coding RNA in the INK4 locus (ANRIL), were lower in patients with AIS compared with control samples, with a prediction value for AIS risk of 0.759, and that ANRIL expression also negatively correlated with the National Institutes of Health Stroke Scale (NIHSS) score. In addition, using a lncRNA microarray, Deng et al (8) screened a three-lncRNA signature [long intergenic non-coding RNA (linc)-DHFRL1-4, small nucleolar RNA host gene 15, and family with sequence similarity 98, member A)], and discovered that this lncRNA-based combination index distinguished patients with AIS from healthy controls, with an AUC >0.84. However, circulating AIS-related lncRNAs still require further investigation.
Accumulating evidence indicates that lncRNAs may be involved in the development of diseases by acting as competing endogenous RNAs (ceRNAs) for microRNA (miRNAs), which are non-coding RNAs of ~20 nucleotides in length that negatively regulate target gene expression at the post-transcriptional level (9). Thus, investigating the lncRNA-related ceRNA axis may provide novel biomarker candidates for AIS; for example, the recent discoveries of ENST00000568297, ENST00000568243, NR_046084, CCAAT/enhancer binding protein, a antisense (AS) RNA 1, lincRNA 884, and matrilin-1-AS RNA 1 (10,11). However, the interacting miRNAs and protein-coding mRNAs in these two studies are not differentially expressed. Therefore, an additional differentially expressed lncRNAs (DELs)/miRNAs (DEMs)/genes (DEGs) network should be constructed to identify potential AIS-associated ceRNA axes. In the present study, mRNA, lncRNA and miRNA expression profiles from AIS samples and healthy samples were collected from the Gene Expression Omnibus (GEO) repository, and were used to establish lncRNA-related ceRNA networks in AIS. The interactions amongst lncRNAs, miRNAs, and genes may provide a novel and valuable insight into understanding the pathogenesis of AIS, and may present novel biomarkers, or targets, for the diagnosis and treatment of AIS.
Materials and methods
Data collection
Five high-throughput datasets, including 4 miRNA (GSE110993, GSE95204, GSE86291 and GSE55937) and 1 mRNA (GSE16561) expression profile, of AIS were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo). Twenty patients with AIS and 20 matched healthy controls were collected from the GSE110993 dataset, and the circulating miRNAs were examined by high-throughput sequencing using Illumina HiScanSQ (Illumina, Inc.; platform, GPL15456) (12). Three patients with AIS and three matched healthy controls were obtained from the GEO database dataset, GSE95204, to detect plasma miRNA expression profiles using the Exiqon miRCURY LNA™ microRNA array system (miRBase; condensed Probe_ID version; version 18; Exiqon, Qiagen). Plasma samples from seven patients with AIS and four matched healthy controls were obtained from the GSE86291 dataset to screen for miRNA biomarkers using the Agilent-046064 Unrestricted_Human_miRNA_V19.0_Microarray (accession no. GPL18402) (13). Using the GSE55937 dataset, miRNA expression was determined in the peripheral blood of 24 patients with AIS and 24 controls using the Affymetrix Multispecies miRNA-3 Array (Affymetrix; Thermo Fisher Scientific, Inc.; accession no. GPL16384) (14). Using the GSE16561 dataset, mRNA and lncRNA expression profiles were analyzed in the peripheral whole blood of 39 patients with AIS and 24 matched healthy controls using the Illumina HumanRef-8 version 3.0 expression beadchip (Illumina, Inc.; platform, GPL6883).
Dataset preprocessing and differential expression analysis
The raw data were downloaded from the GEO database. For the microarray datasets, the data were quantile-normalized, and the DEGs, DELs and DEMs were identified using the linear models for microarray data (limma) software package (version 3.34.0; http://www.bioconductor.org/packages/release/bioc/html/limma.html) (15) in R Bioconductor (version 3.4.1; http://www.R-project.org). To sequence the dataset, fastp software (version 1.0; http://github.com/OpenGene/fastp) was used to perform the adapter trimming and quality control (16), and the DEMs were subsequently screened using DESeq2 (version 1.16.1; http://bioconductor.org/packages/3.5/bioc/html/DESeq2.html) (17). P<0.05 was defined as the statistical threshold for DEMs to obtain overlap among different datasets. P<0.05 and log2fold change (FC) >0.33 were defined as the statistical cut-off values for DEGs and DELs. A Venn diagram was created to determine DEMs shared between any two datasets using Draw Venn Diagram online tool (version 1.0; http://bioinformatics.psb.ugent.be/webtools/Venn). Hierarchical clustering was performed using the pheatmap package (version 1.0.8; http://cran.r-project.org/web/packages/pheatmap) to determine the intersection between differentially expressed RNAs in different samples.
Functional enrichment analysis
The clusterProfiler tool (version 3.2.11; http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) was used to analyze the potential functions of DEGs, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Adjusted P<0.05 was set as the criterion.
Protein-protein interaction (PPI) network
PPI data of DEGs were collected from the Search Tool for the Retrieval of Interacting Genes (STRING) database (version 10.0; http://string-db.org) (18). Interactions with a combined score of >700 were used to construct the PPI network. Several topological features of the nodes (proteins) in the PPI network were calculated using the CytoNCA plugin (version 2.1.6; http://apps.cytoscape.org/apps/cytonca) (19) in Cytoscape software (version 3.0; http://cytoscape.org), to identify hub genes, including degree, betweenness and closeness centrality. The Molecular Complex Detection (MCODE; version 1.4.2; http://apps.cytoscape.org/apps/mcode) plugin of the Cytoscape software was used to extract highly interconnected sub-networks, with a MCODE score of >6 and number of nodes >6 set as the threshold values (20).
Determining the overall ceRNA regulatory network
The miRcode (version 1.0; http://www.mircode.org) (21), starBase (version 2.0; http://starbase.sysu.edu.cn/starbase2) (22), DIANA-LncBase (version 2.0; http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted) (23), and miRwalk (version 2.0; zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2) (24) databases were used to predict the miRNAs interacting with DELs, which were subsequently intersected with the DEMs shared between two datasets to obtain DEL/DEM interaction pairs. The target genes of the crosstalk between the DEMs and DEGs were predicted using the miRwalk database (24). The DEL/DEM and DEM/DEG interactors were subsequently linked to construct the ceRNA network using the Cytoscape software.
Determining potential AIS-relevant ceRNA networks
The AIS-related KEGG pathways were downloaded from the Comparative Toxicogenomics Database (CTD; version 1.0; http://ctdbase.org) and were subsequently overlapped with the pathways enriched by the DEGs in the overall ceRNA network. The ceRNA network, including the pathway-related DEGs, was extracted individually to define a potential AIS-related ceRNA network. Known lncRNAs were collected from the LncRNADisease database (version 2.0; http://www.cuilab.cn/lncrnadisease) to determine whether the DELs identified in the AIS-related ceRNA network were novel.
Results
DEGs and DELs analyses
Based on the microarray data analysis between patients with AIS and healthy controls using the limma method, 2,041 DEGs were screened from the GSE16561 dataset. A total of 1,337 DEGs were found to be upregulated, such as paxillin (PXN), ras homolog family member A (RHOA), STAT1, and growth factor receptor bound protein 2 (GRB2), and 704 downregulated, including FYN proto-oncogene, Src family tyrosine kinase (FYN) (Table I). In addition, two upregulated DELs [human leukocyte antigen complex group 27 (HCG27), and inositol-tetrakisphosphate 1-kinase AS RNA 1 (ITPK1-AS1)] and three downregulated DELs [minichromosome maintenance complex component 3 associated protein-antisense RNA 1 (MCM3AP-AS1), leucine rich repeat containing 37 member A4 (LRRC37A4P), and lincRNA 1089 (LINC01089)] were identified (Table I). The heatmap revealed that the differentially expressed RNAs identified from this dataset could distinguish the AIS samples from the healthy controls (Fig. 1).
Table I.
Differentially expressed RNAs identified in patients with acute ischemic stroke from datasets in the Gene Omnibus database repository.
| Dataset accession number | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GSE110993 | GSE95204 | GSE86291 | GSE55937 | GSE16561 | ||||||
| RNA type | Log2FC | P-value | Log2FC | P-value | Log2FC | P-value | Log2FC | P-value | Log2FC | P-value |
| MicroRNA | ||||||||||
| hsa-miR-99b | 1.06 | 2.14×10−6 | 0.77 | 1.69×10−3 | ||||||
| hsa-miR-551a | 0.43 | 3.40×10−2 | 0.15 | 2.13×10−2 | ||||||
| hsa-miR-1263 | 1.84 | 3.57×10−3 | 0.42 | 3.95×10−2 | ||||||
| hsa-miR-125a | 1.30 | 8.51×10−8 | 0.38 | 4.60×10−2 | ||||||
| hsa-miR-1283 | 1.57 | 2.59×10−2 | 3.38 | 3.94×10−3 | ||||||
| hsa-miR-19a | −1.00 | 9.96×10−3 | −0.54 | 3.55×10−2 | ||||||
| hsa-let-7i | −1.05 | 4.97×10−4 | −0.54 | 3.96×10−2 | ||||||
| hsa-miR-345-5p | −0.67 | 2.53×10−2 | −0.35 | 1.66×10−2 | ||||||
| hsa-miR-5100 | −0.85 | 3.54×10−2 | −1.87 | 6.54×10−3 | ||||||
| hsa-miR-4667-5p | −1.81 | 3.73×10−2 | −0.41 | 2.74×10−2 | ||||||
| Long non-coding RNA | ||||||||||
| HCG27 | 0.52 | 2.41×10−4 | ||||||||
| ITPK1-AS1 | 0.39 | 3.71×10−2 | ||||||||
| MCM3AP-AS1 | −0.43 | 6.69×10−8 | ||||||||
| LRRC37A4P | −0.38 | 1.25×10−3 | ||||||||
| LINC01089 | −0.34 | 6.05×10−5 | ||||||||
| mRNA | ||||||||||
| PXN | 0.46 | 3.15×10−4 | ||||||||
| FYN | −0.53 | 1.19×10−5 | ||||||||
| RHOA | 0.47 | 2.66×10−6 | ||||||||
| STAT1 | 0.54 | 1.14×10−4 | ||||||||
| GRB2 | 0.38 | 6.20×10−3 | ||||||||
FC, fold change; miR, microRNA; ITPK1-AS1, inositol-tetrakisphosphate 1-kinase-antisense RNA 1; HCG27, human leukocyte antigen complex group 27; MCM3AP-AS1, minichromosome maintenance complex component 3 associated protein-antisense RNA 1; LRRC37A4P, leucine rich repeat containing 37 member A4; LINC01089, long intergenic non-protein coding RNA 1089; PXN, paxillin; FYN, FYN Proto-oncogene tyrosine-protein kinase; RHOA, Ras homolog family member A; GRB2, growth factor receptor bound protein 2.
Figure 1.
Hierarchical clustering heat map for the differentially expressed mRNAs in acute ischemic stroke. Data were obtained from datasets in the Gene Expression Omnibus database. On the horizontal axis, the white box indicates the control samples, and the black box represents the acute ischemic stroke samples. The vertical axis indicates the gene expression level. Red, high expression; blue, low expression.
DEMs analysis
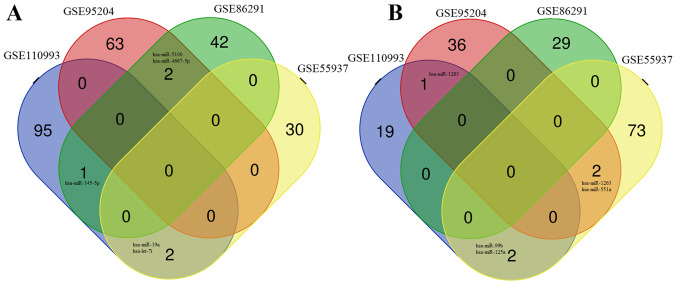
The microarray data analysis between AIS samples and healthy controls revealed 104 DEMs in the GSE95204 dataset (39 upregulated; 65 downregulated), 74 DEMs in the GSE86291 dataset (29 upregulated; 45 downregulated), and 111 DEMs in the GSE55937 dataset (77 upregulated; 32 downregulated) (Table I). Sequence analysis between AIS samples and healthy controls using the DESeq2 method identified 120 DEMs in the GSE110993 dataset, 22 that were upregulated, and 98 that were downregulated. Venn diagram analysis demonstrated that 10 DEMs were shared between at least two of the four miRNA expression profile datasets (Table I), including: Five downregulated miRNAs; hsa-miRNA (miR)-345-5p, hsa-miR-19a, hsa-let-7i, hsa-miR-5100 and hsa-miR-4667-5p (Fig. 2A); and five upregulated miRNAs; hsa-miR-1283, hsa-miR-99b, hsa-miR-125a, hsa-miR-1263 and hsa-miR-551a (Fig. 2B).
Figure 2.
Venn diagram of the common DEMs. DEMs were identified in at least two expression profile datasets out of four for miRNAs in acute ischemic stroke (accession nos. GSE95204, GSE86291, GSE55937, and GSE110993), and were either (A) downregulated or (B) upregulated. DEM, differentially expressed microRNA.
Functional enrichment analysis of the DEGs
Functional enrichment analysis of all the DEGs revealed that the DEGs were enriched in 921 GO biological processes and 56 significant KEGG pathways. Among them, inflammatory-related functions, such as ‘I-κB kinase/NF-κB signaling’/‘NF-κB signaling pathway’, ‘T cell differentiation’/‘Th17 cell differentiation’/‘Th1 and Th2 cell differentiation’, ‘T cell activation’/‘T cell receptor signaling pathway’, were the most significant, and were identified by both GO and KEGG enrichment analyses (Fig. 3). Thus, this indicated that inflammatory genes may be crucial for AIS development.
Figure 3.
Functional enrichment analysis for the differentially expressed genes. (A) Top 30 significant GO terms. (B) Top 10 significant KEGG pathways. KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, gene ontology.
PPI network of DEGs
Using the STRING database, a total of 6,246 PPI interaction pairs were predicted from the 1,235 DEGs used to construct the PPI network (data not shown). Twenty genes were identified as hub genes because they ranked in the top 60 (5% of all DEGs), according to the ranking results of all three topological features (Table II). Furthermore, seven significant modules were extracted from the PPI network, in which the hub genes, PXN, FYN, RHOA, STAT1 and GRB2 were enriched in module 7 (Fig. 4; Table III). Functional analysis revealed that the genes in module 7 may be involved in AIS by participating in 78 significant KEGG pathways (Table IV), 61 of which included at least one of these five hub genes; T cell receptor signaling pathway (GRB2/FYN/RHOA), chemokine signaling pathway (PXN/GRB2/STAT1/RHOA), focal adhesion (PXN/GRB2/FYN/RHOA) and Th17 cell differentiation (STAT1). These findings suggested that these five inflammatory genes may be important for AIS.
Table II.
Centralities analysis of the top 20 hub genes identified in the protein-protein interaction network from differentially expressed genes in acute ischemic stress.
| Gene | Degree centrality | Gene | Betweenness centrality | Gene | Closeness centrality |
|---|---|---|---|---|---|
| STAT3 | 96 | HDAC1 | 0.066077 | STAT3 | 0.39361 |
| RHOA | 89 | BCL2 | 0.065222 | MAPK14 | 0.384987 |
| FYN | 85 | STAT3 | 0.062793 | BCL2 | 0.383381 |
| MAPK14 | 80 | RHOA | 0.056532 | CREBBP | 0.38289 |
| CREBBP | 79 | ACTB | 0.050714 | FOS | 0.381667 |
| BCL2 | 72 | CREBBP | 0.049029 | RHOA | 0.380331 |
| CDC42 | 71 | CDC42 | 0.047746 | MAPK1 | 0.377329 |
| HDAC1 | 71 | MAPK14 | 0.041837 | STAT1 | 0.376734 |
| STAT1 | 71 | FYN | 0.040107 | MAPK3 | 0.375196 |
| MAPK1 | 71 | STAT1 | 0.036472 | FYN | 0.369855 |
| FOS | 71 | NOTCH1 | 0.035509 | NOTCH1 | 0.36724 |
| MAPK3 | 66 | FOS | 0.034142 | ACTB | 0.366564 |
| GRB2 | 61 | HIF1A | 0.033411 | HDAC1 | 0.363443 |
| ACTB | 58 | GRB2 | 0.030588 | PTEN | 0.363332 |
| NOTCH1 | 53 | MAPK1 | 0.029564 | CDC42 | 0.362891 |
| TLR4 | 49 | GSK3B | 0.023502 | GSK3B | 0.362891 |
| PXN | 49 | PTEN | 0.023476 | PXN | 0.362121 |
| GSK3B | 47 | MAPK3 | 0.020408 | TLR4 | 0.360265 |
| HIF1A | 47 | TLR4 | 0.019773 | GRB2 | 0.360048 |
| PTEN | 47 | PXN | 0.013521 | HIF1A | 0.35929 |
STAT3, signal transducer and activator of transcription 3; RHOA, Ras homolog family member A; FYN, FYN Proto-oncogene tyrosine-protein kinase; MAPK14, mitogen-activated protein kinase 14; CREBBP, CREB binding protein; BCL2, BCL2 apoptosis regulator; CDC42, cell division cycle 42; HDAC1, histone deacetylase 1; STAT1, signal transducer and activator of transcription 1; MAPK1, mitogen-activated protein kinase 1; FOS, Fos proto-oncogene AP-1 transcription factor subunit; MAPK3, mitogen-activated protein kinase 3; GRB2, growth factor receptor bound protein 2; ACTB, actin β; NOTCH1, notch receptor 1; TLR4, toll-like receptor 4; PXN, paxillin; GSK3B, glycogen synthase kinase 3 β; HIF1A, hypoxia inducible factor 1 subunit α; PTEN, phosphatase and tensin homolog.
Figure 4.
A significant module that includes hub genes identified in the protein and protein interaction network for the differentially expressed genes in acute ischemic stroke. Blue, downregulated; red, upregulated. PXN, paxillin; RAF1, Raf-1 proto-oncogene serine/threonine kinase; TLR2, toll-like receptor 2; RPS6KA5, ribosomal protein S6 kinase A5; ITGA2B, integrin subunit α 2b; PRKCQ, protein kinase C θ; IL-1B, interleukin 1β; CBL, Cbl proto-oncogene; ZAP70, ζ chain of T cell receptor-associated protein kinase 70; TPM1, tropomyosin 1; PAK1, p21 (RAC1) activated kinase 1; RHOC, ras homolog family member C; ITGB5, integrin subunit β5; PAK2, p21 (RAC1) activated kinase 2; GRB2, growth factor receptor bound protein 2; PIK3AP1, phosphoinositide-3-kinase adaptor protein 1; FYN, FYN Proto-oncogene tyrosine-protein kinase; TPM2, tropomyosin 2; STAT1, signal transducer and activator of transcription 1; PTGS2, prostaglandin-endoperoxide synthase 2; FGR, FGR proto-oncogene Src family tyrosine kinase; ITPR3, inositol 1,4,5-trisphosphate receptor type 3; SYK, spleen-associated tyrosine kinase; HCK, HCK proto-oncogene Src family tyrosine kinase; RPS6KA3, ribosomal protein S6 kinase A3; CD79B, CD79b molecule; MAP4K1, mitogen-activated protein kinase kinase kinase kinase 1; CREB1, cAMP responsive element binding protein 1; SLA, Src-like adaptor; ITK, IL2 inducible T cell kinase; RHOA, Ras homolog family member A.
Table III.
Significant modules extracted from the protein-protein interaction network of differentially expressed genes in acute ischemic stress.
| Module | Molecular complex detection score | Number of nodes | Number of edges | Genes |
|---|---|---|---|---|
| 1 | 31.632 | 39 | 601 | RPS5, EIF3D, SSR1, RPL36, EIF3G, EIF4B, RPL8, CASC3, SRP14, SSR2, RPS23, RNPS1, RPS9, RPL13, MRPS12, MRPL11, EIF3F, RPL4, RPS15A, IMP3, RPLP0, RPS2, RPL3, RPL9, RPSA, RPL22, UPF2, ETF1, MRPL24, RPL12, EIF3A, RPS4X, RPL10A, UPF3A, RPL13A, SRPRB, RPS3, RPL17, RPS15 |
| 2 | 26.000 | 26 | 325 | HEBP1, CCR7, ANXA1, APP, CXCR5, CCL5, CXCL16, CXCR1, PPBP, CCR1, PNOC, C3AR1, FPR1, S1PR1, CXCR2, P2RY13, LPAR5, S1PR5, FPR2, GPR18, C5AR1, S1PR3, RGS18, GNAI3, LPAR2, ADRA2C |
| 3 | 12.308 | 14 | 80 | OASL, IFI27, STAT2, EEF1G, MX2, RNASEL, ADAR, GBP2, IFIT1, IFIT3, IFIT2, MX1, IFITM3, OAS1 |
| 4 | 10.000 | 10 | 45 | NDUFB7, NDUFB3, NDUFB2, NDUFB11, NDUFB8, NDUFS8, NDUFV1, NDUFA12, NDUFS5, NDUFA11 |
| 5 | 8.286 | 36 | 145 | P2RY10, RGS2, TBL1X, MED10, CDK4, VWF, SMARCD3, F13A1, GPR65, SERPING1, MED13L, CD8A, PROK2, F2RL1, CD3D, PTAFR, CEBPB, LTB4R, CD36, VEGFB, NCOA1, CD8B, SERPINA1, HLA-DRB1, CD3E, CD247, F5, CDK19, NCOA3, KLF4, PROS1, ACTN1, CEBPD, HELZ2, CEBPA, CD3G |
| 6 | 8.000 | 8 | 28 | STAG2, B9D2, TAOK1, RAD21, CLIP1, NDEL1, AHCTF1, MIS12 |
| 7 | 7.677 | 32 | 119 | PXN, RAF1, TLR2, RPS6KA5, ITGA2B, PRKCQ, IL1B, CBL, ZAP70, TPM1, PAK1, RHOC, ITGB5, PAK2, GRB2, PIK3AP1, FYN, TPM2, STAT1, PTGS2, FGR, ITPR3, SYK, HCK, RPS6KA3, CD79B, MAP4K1, CREB1, SLA, ITK, RHOA, CD19 |
Table IV.
KEGG pathway enrichment analysis for module 7 identified from the protein-protein interaction networks of differentially expressed genes in acute ischemic stroke.
| KEGG ID | Description | P-value adjusted | Gene |
|---|---|---|---|
| hsa04660 | T cell receptor signaling pathway | 1.29×10−8 | RAF1/PRKCQ/ZAP70/PAK1/PAK2/GRB2/FYN/ITK/RHOA |
| hsa04625 | C-type lectin receptor signaling pathway | 2.78×10−7 | RAF1/IL1B/PAK1/STAT1/PTGS2/ITPR3/SYK/RHOA |
| hsa04062 | Chemokine signaling pathway | 1.19×10−6 | PXN/RAF1/PAK1/GRB2/STAT1/FGR/HCK/ITK/RHOA |
| hsa04510 | Focal adhesion | 1.19×10−6 | PXN/RAF1/ITGA2B/PAK1/ITGB5/PAK2/GRB2/FYN/RHOA |
| hsa05205 | Proteoglycans in cancer | 1.19×10−6 | PXN/RAF1/TLR2/CBL/PAK1/ITGB5/GRB2/ITPR3/RHOA |
| hsa04662 | B cell receptor signaling pathway | 6.14×10−6 | RAF1/GRB2/PIK3AP1/SYK/CD79B/CD19 |
| hsa05163 | Human cytomegalovirus infection | 3.26×10−5 | PXN/RAF1/IL1B/GRB2/PTGS2/ITPR3/CREB1/RHOA |
| hsa05152 | Tuberculosis | 7.77×10−5 | RAF1/TLR2/IL1B/STAT1/SYK/CREB1/RHOA |
| hsa04151 | PI3K-AKT signaling pathway | 7.98×10−5 | RAF1/TLR2/ITGA2B/ITGB5/GRB2/PIK3AP1/SYK/CREB1/CD19 |
| hsa05167 | Kaposi sarcoma-associated herpes virus infection | 8.02×10−5 | RAF1/STAT1/PTGS2/ITPR3/SYK/HCK/CREB1 |
| hsa04380 | Osteoclast differentiation | 1.09×10−4 | IL1B/GRB2/FYN/STAT1/SYK/CREB1 |
| hsa04650 | Natural killer cell mediated cytotoxicity | 1.14×10−4 | RAF1/ZAP70/PAK1/GRB2/FYN/SYK |
| hsa04010 | Mitogen activated protein kinase signaling pathway | 1.32×10−4 | RAF1/RPS6KA5/IL1B/PAK1/PAK2/GRB2/RPS6KA3/MAP4K1 |
| hsa04810 | Regulation of actin cytoskeleton | 1.44×10−4 | PXN/RAF1/ITGA2B/PAK1/ITGB5/PAK2/RHOA |
| hsa04012 | ErbB signaling pathway | 1.69×10−4 | RAF1/CBL/PAK1/PAK2/GRB2 |
| hsa05165 | Human papillomavirus infection | 2.43×10−4 | PXN/RAF1/ITGA2B/ITGB5/GRB2/STAT1/PTGS2/CREB1 |
| hsa04722 | Neurotrophin signaling pathway | 7.17×10−4 | RAF1/RPS6KA5/GRB2/RPS6KA3/RHOA |
| hsa04611 | Platelet activation | 8.26×10−4 | ITGA2B/FYN/ITPR3/SYK/RHOA |
| hsa04664 | Fc ε RI signaling pathway | 9.55×10−4 | RAF1/GRB2/FYN/SYK |
| hsa05170 | Human immunodeficiency virus 1 infection | 9.55×10−4 | PXN/RAF1/TLR2/PAK1/PAK2/ITPR3 |
| hsa05211 | Renal cell carcinoma | 9.55×10−4 | RAF1/PAK1/PAK2/GRB2 |
| hsa05140 | Leishmaniasis | 1.20×10−3 | TLR2/IL1B/STAT1/PTGS2 |
| hsa04014 | Ras signaling pathway | 1.42×10−3 | RAF1/ZAP70/PAK1/PAK2/GRB2/RHOA |
| hsa04072 | Phospholipase D signaling pathway | 1.45×10−3 | RAF1/GRB2/FYN/SYK/RHOA |
| hsa05161 | Hepatitis B | 2.10×10−3 | RAF1/TLR2/GRB2/STAT1/CREB1 |
| hsa04360 | Axon guidance | 3.06×10−3 | RAF1/PAK1/PAK2/FYN/RHOA |
| hsa04928 | Parathyroid hormone synthesis, secretion and action | 3.53×10−3 | RAF1/ITPR3/CREB1/RHOA |
| hsa04659 | Type 17 T helper 7 cell differentiation | 3.54×10−3 | PRKCQ/IL1B/ZAP70/STAT1 |
| hsa05206 | MicroRNAs in cancer | 3.90×10−3 | RAF1/RPS6KA5/TPM1/GRB2/PTGS2/RHOA |
| hsa05203 | Viral carcinogenesis | 4.22×10−3 | PXN/GRB2/SYK/CREB1/RHOA |
| hsa04370 | Vascular endothelial growth factor signaling pathway | 6.55×10−3 | PXN/RAF1/PTGS2 |
| hsa04270 | Vascular smooth muscle contraction | 6.71×10−3 | RAF1/PRKCQ/ITPR3/RHOA |
| hsa04915 | Estrogen signaling pathway | 7.70×10−3 | RAF1/GRB2/ITPR3/CREB1 |
| hsa05321 | Inflammatory bowel disease | 8.00×10−3 | TLR2/IL1B/STAT1 |
| hsa04917 | Prolactin signaling pathway | 9.19×10−3 | RAF1/GRB2/STAT1 |
| hsa04150 | mTOR signaling pathway | 9.73×10−3 | RAF1/GRB2/RPS6KA3/RHOA |
| hsa04921 | Oxytocin signaling pathway | 9.73×10−3 | RAF1/PTGS2/ITPR3/RHOA |
| hsa05100 | Bacterial invasion of epithelial cells | 4.16×10−2 | PXN/CBL/RHOA |
| hsa05220 | Chronic myeloid leukemia | 1.06×10−2 | RAF1/CBL/GRB2 |
| hsa04022 | Cyclic GMP-Protein kinase G signaling pathway | 1.22×10−2 | RAF1/ITPR3/CREB1/RHOA |
| hsa05210 | Colorectal cancer | 1.44×10−2 | RAF1/GRB2/RHOA |
| hsa04621 | Nucleotide-binding oligomerization domain-like receptor signaling pathway | 1.48×10−2 | IL1B/STAT1/ITPR3/RHOA |
| hsa04540 | Gap junction | 1.48×10−2 | RAF1/GRB2/ITPR3 |
| hsa04658 | Type 1 and 2 T helper cell differentiation | 1.64×10−2 | PRKCQ/ZAP70/STAT1 |
| hsa04912 | Gonadotropin-releasing hormone signaling pathway | 1.66×10−2 | RAF1/GRB2/ITPR3 |
| hsa05215 | Prostate cancer | 1.76×10−2 | RAF1/GRB2/CREB1 |
| hsa05169 | Epstein-Barr virus infection | 1.98×10−2 | TLR2/STAT1/SYK/CD19 |
| hsa04620 | Toll-like receptor signaling pathway | 2.03×10−2 | TLR2/IL1B/STAT1 |
| hsa05020 | Prion diseases | 2.18×10−2 | IL1B/FYN |
| hsa04024 | Cyclic AMP signaling pathway | 2.23×10−2 | RAF1/PAK1/CREB1/RHOA |
| hsa04670 | Leukocyte transendothelial migration | 2.26×10−2 | PXN/ITK/RHOA |
| hsa04725 | Cholinergic synapse | 2.26×10−2 | FYN/ITPR3/CREB1 |
| hsa04071 | Sphingolipid signaling pathway | 2.58×10−2 | RAF1/FYN/RHOA |
| hsa04926 | Relaxin signaling pathway | 3.18×10−2 | RAF1/GRB2/CREB1 |
| hsa04910 | Insulin signaling pathway | 3.56×10−2 | RAF1/CBL/GRB2 |
| hsa05162 | Measles | 3.58×10−2 | TLR2/IL1B/STAT1 |
| hsa05418 | Fluid shear stress and atherosclerosis | 3.59×10−2 | ITGA2B/IL1B/RHOA |
| hsa05130 | Pathogenic Escherichia coli infection | 4.16×10−2 | FYN/RHOA |
| hsa05213 | Endometrial cancer | 4.53×10−2 | RAF1/GRB2 |
| hsa05160 | Hepatitis C | 4.54×10−2 | RAF1/GRB2/STAT1 |
| hsa04630 | Janus kinase-STAT signaling pathway | 4.97×10−2 | RAF1/GRB2/STAT1 |
KEGG, Kyoto Encyclopedia for Genes and Genomes.
Construction and functional analysis of the ceRNA network
Based on four miRNA databases (miRcode, starBase, DIANA-LncBase and miRwalk), 9 DEMs (hsa-let-7i, hsa-miR-125a, hsa-miR-1283, hsa-miR-19a, hsa-miR-345-5p, hsa-miR-4667-5p, hsa-miR-5100, hsa-miR-551a and hsa-miR-99b), were predicted to interact with 4 DELs (ITPK1-AS1, LINC01089, MCM3AP-AS1 and HCG27), which constituted 29 interaction pairs. Subsequently, 7,995 regulatory pairs between 9 DEMs, and 712 DEGs were detected in the miRwalk database, which was used for constructing the ceRNA network following integration with the DEM-DEL interacting relationships (data not shown).
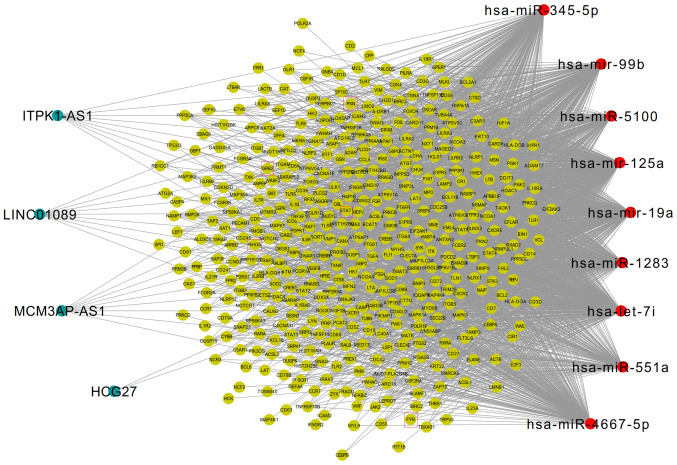
Furthermore, the mRNAs in the ceRNA regulatory network were enriched in 66 KEGG signaling pathways, and 53 overlapped with the 191 AIS-associated KEGG pathways collected from the CTD database. The 436 genes enriched in these 53 KEGG pathways were subsequently extracted to construct a potential AIS-related ceRNA network (Fig. 5). In this network, the five inflammatory genes (PXN, FYN, STAT1, RHOA, GRB2) were included. Downregulated FYN was predicted to be regulated by upregulated hsa-miR-125a, whereas downregulated MCM3AP-AS1 and LINC01089 could also interact with hsa-miR-125a, thus forming the MCM3AP-AS1/LINC01089/hsa-miR-125a/FYN ceRNA axis (Fig. 6). Furthermore, the upregulated expression of RHOA, GRB2 and STAT1 was predicted to be regulated by the downregulated expression of hsa-let-7i, and this miRNA could also interact with the upregulated lncRNA, ITPK1-AS1. Accordingly, the ITPK1-AS1/hsa-let-7i/RHOA/GRB2/STAT1 ceRNA axes may also be important for AIS. Additionally, the upregulated expressions of PXN and HCG27 could interact with hsa-miR-19a to establish the HCG27/hsa-miR-19a/PXN ceRNA axis for AIS (Fig. 6). Notably, to the best of our knowledge this is the first time that the LncRNADisease database has shown that the lncRNAs involved in these ceRNA networks are associated with AIS, which suggested that they may be newly identified targets for the diagnosis and treatment of AIS. These five genes participated in 34 significant KEGG pathways (Table V), 26 of which were similar to the results of module 7, including Th17 cell differentiation (STAT1), T cell receptor signaling pathway (FYN/GRB2/RHOA), chemokine signaling pathway (GRB2/RHOA/STAT1/PXN), and focal adhesion (FYN/GRB2/RHOA/PXN). Therefore, the lncRNA ceRNA axes associated with these five inflammatory DEGs may be of interest.
Figure 5.
Competing endogenous RNAs interaction network of all potential acute ischemic stroke-related long noncoding RNA/miRNA/mRNA axes. Blue, long noncoding RNAs; green, mRNA; red, microRNA. Hub genes are presented in red boxes. miR/miRNA, microRNA; PXN, paxillin; GRB2, growth factor receptor bound protein 2; FYN, FYN Proto-oncogene tyrosine-protein kinase; STAT1, signal transducer and activator of transcription 1; RHOA, Ras homolog family member A.
Figure 6.
Competing endogenous RNA interaction network between five inflammatory differentially expressed hub genes, and their related long noncoding RNA/miRNA axes in AIS. Purple, hub genes; red, microRNAs; green, long non-coding RNAs. miR/miRNA, microRNA; PXN, paxillin; GRB2, growth factor receptor bound protein 2; FYN, FYN Proto-oncogene tyrosine-protein kinase; STAT1, signal transducer and activator of transcription 1; RHOA, Ras homolog family member A.
Table V.
KEGG pathways enriched with potential crucial genes in the AIS-relevant ceRNA network.
| KEGG ID | Description | P-value adjusted | Gene |
|---|---|---|---|
| hsa05152 | Tuberculosis | 2.15×10−9 | RHOA/STAT1 |
| hsa04659 | Th17 cell differentiation | 6.91×10−8 | STAT1 |
| hsa04380 | Osteoclast differentiation | 6.91×10−8 | FYN/GRB2/STAT1 |
| hsa05140 | Leishmaniasis | 2.94×10−7 | STAT1 |
| hsa04660 | T cell receptor signaling pathway | 4.90×10−7 | FYN/GRB2/RHOA |
| hsa04658 | Type 1 and 2 T helper cell differentiation | 4.90×10−7 | STAT1 |
| hsa05164 | Influenza A | 3.10×10−6 | STAT1 |
| hsa04662 | B cell receptor signaling pathway | 1.84×10−5 | GRB2 |
| hsa04621 | Nucleotide-binding oligomerization domain-like receptor signaling pathway | 3.66×10−5 | RHOA/STAT1 |
| hsa05223 | Non-small cell lung cancer | 2.35×10−4 | GRB2 |
| hsa05169 | Epstein-Barr virus infection | 5.75×10−4 | STAT1 |
| hsa04611 | Platelet activation | 5.87×10−4 | FYN/RHOA |
| hsa05145 | Toxoplasmosis | 6.37×10−4 | STAT1 |
| hsa04062 | Chemokine signaling pathway | 6.37×10−4 | GRB2/RHOA/STAT1/PXN |
| hsa05162 | Measles | 6.37×10−4 | STAT1 |
| hsa04068 | FoxO signaling pathway | 6.38×10−4 | GRB2 |
| hsa05321 | Inflammatory bowel disease | 1.32×10−3 | STAT1 |
| hsa05221 | Acute myeloid leukemia | 1.58×10−3 | GRB2 |
| hsa04670 | Leukocyte transendothelial migration | 2.76×10−3 | RHOA/PXN |
| hsa04071 | Sphingolipid signaling pathway | 2.91×10−3 | FYN/RHOA |
| hsa04919 | Thyroid hormone signaling pathway | 4.31×10−3 | STAT1 |
| hsa04650 | Natural killer cell mediated cytotoxicity | 5.16×10−3 | FYN/GRB2 |
| hsa04664 | Fc e RI signaling pathway | 5.21×10−3 | FYN/GRB2 |
| hsa05133 | Pertussis | 7.11×10−3 | RHOA |
| hsa05160 | Hepatitis C | 7.12×10−3 | GRB2/STAT1 |
| hsa05203 | Viral carcinogenesis | 9.56×10−3 | GRB2/RHOA/PXN |
| hsa04722 | Neurotrophin signaling pathway | 1.11×10−2 | GRB2/RHOA |
| hsa04010 | Mitogen activated protein kinase signaling pathway | 1.32×10−2 | GRB2 |
| hsa05161 | Hepatitis B | 1.35×10−2 | GRB2/STAT1 |
| hsa05205 | Proteoglycans in cancer | 1.56×10−2 | GRB2/RHOA |
| hsa04370 | Vascular endothelial growth factor signaling pathway | 1.85×10−2 | PXN |
| hsa04510 | Focal adhesion | 2.22×10−2 | FYN/GRB2/RHOA/PXN |
| hsa05212 | Pancreatic cancer | 2.82×10−2 | STAT1 |
| hsa04915 | Estrogen signaling pathway | 3.12×10−2 | GRB2 |
KEGG, Kyoto Encyclopedia for Genes and Genomes.
Discussion
Extensive evidence indicates that AIS is commonly associated with the activation of immune-inflammatory responses, which leads to the increased expression of various pro-inflammatory cytokines, such as interleukin (IL)-6, IL-1 and C-reactive protein, and, subsequently, neuronal cell death (25,26). It has been revealed through several in vitro and in vivo studies that lncRNAs involved in the inflammatory process may be potential targets for the diagnosis and treatment of AIS (27–31). For example, Zhang et al (28) discovered that lncRNA-1810034E14Rik is significantly decreased in oxygen-glucose deprivation-induced microglial cells and in transient middle cerebral artery occlusion (MCAO) mice, and that 1810034E14Rik overexpression decreases the infarct volume and alleviates the neuronal damage by reducing the expression of pro-inflammatory cytokines IL-1β, tumor necrosis factor (TNF)-α and IL-6. Wang et al (29) reported that levels of the lncRNA H19, increased in the plasma, white blood cells and brain of MCAO mice. Consequently, intracerebroventricular injection of H19 small interfering RNA reduced the infarct volume and brain edema through the suppressed release of pro-inflammatory IL-1β, TNF-α, and increased anti-inflammatory IL-10 secretion (29). Similarly, in vitro experiments demonstrated that H19 knockdown attenuated brain tissue loss and neurological deficits by blocking M1 microglial polarization, and the production of TNF-α. Genetic knockdown of the lncRNAs metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and Maclpil, attenuated inflammatory injuries attained in MCAO mice (30,31), and rare lncRNAs (ANRIL and H19) are associated with inflammation in patients with AIS (7,29,32). In the present study, high-throughput datasets of patients with AIS were used to identify four crucial lncRNAs (MCM3AP-AS1, LINC01089, ITPK1-AS1 and HCG27) that may regulate T cell receptor signaling pathway genes (FYN, GRB2 and RHOA), and inflammatory chemokine signaling pathway genes (GRB2, RHOA, STAT1 and PXN) by functioning as ceRNAs that competitively bind with miRNAs. Previous studies have cited cancer-related functions for MCM3AP-AS1, LINC01089 and ITPK1-AS1 (33–36), but none has associated them with the pathogenesis of AIS, indicating that they may serve as novel, non-invasive biomarkers for the diagnosis and treatment of AIS. However, although HCG27 has been reported to be downregulated in IS (11), the present study found it to be upregulated, which may be attributed to the difference in the sample selection (chronic vs. acute) and the number of patients (3 vs. 39). Thus, the results presented in this study may be more realistic for AIS.
Although no direct evidence was provided in this study to demonstrate that these lncRNAs are involved in AIS, the association with AIS of their interactive miRNA-mRNA pairs indirectly suggests their role. For example, it had been reported that the expression of RhoA is increased in cerebral IS model mice (37), and the use of RhoA inhibitors reduced neuronal cell apoptosis and improved neurobehavioral outcomes (37,38). In patients with stroke, T cells exhibit high RhoA activity, which may mediate the secretion of interferon γ and IL-18 (39). There is also a significant correlation between the amount of inflammation, as indicated by the number of CD11b+/Iba+ cells, and the amount of RhoA activation in nerves from in vivo rat stroke models (40). Microarray analysis reveals an enhanced expression of GRB2 24 at 72 h following ischemia, which has been confirmed by ELISA (41,42). In addition, IS induces STAT1 expression, which is an important mediator of pro-inflammatory cytokines, such as IL-1β and TNF-α, and STAT1 inhibition by silymarin ameliorated inflammation-mediated brain tissue injury (43). Hsa-let-7i is decreased in patients with AIS (14,38), and inversely correlated with NIHSS score at admission and infarct volume (44,45). In vitro functional analysis demonstrated that hsa-let-7i might be involved in AIS by negatively regulating inflammatory cluster of differentiation (CD)86 signaling in T helper cells, and IL-8 signaling pathways (45). Thus, ITPK1-ASI may be upregulated in AIS to further sequester hsa-let-7i and prevent it from inhibiting the expression of RHOA/GRB2/STAT-1, ultimately resulting in the upregulation of RHOA/GRB2/STAT1 in AIS.
Using a MCAO AIS model, Franciska et al (46) revealed that PXN expression is upregulated, especially following 1 h of ischemia (46), and jasminoidin and ursodeoxycholic acid may be effective treatments of focal cerebral ischemia-reperfusion injury through downregulating PXN (47). Hsa-miR-19a is decreased in patients with AIS compared with control samples (14), especially within small vessel stroke (48), and this downregulated expression is hypothesized to serve a crucial role in the pathogenesis of stroke by triggering the increased expression of CD46, an important transmembrane protein that induces inflammation (49). Thus, HCG27 may also be upregulated to sponge hsa-miR-19a, thus facilitating the upregulation of PXN by reducing the expression of hsa-miR-19a, and subsequently promoting inflammation, and the development of AIS.
Experimental studies have demonstrated that the genetic knockdown of FYN kinases may have a potential neuroprotective effect for IS by decreasing neuronal apoptosis, cerebral edema, and enhancing the rate of neurological recovery (50,51). These findings suggest that FYN may be upregulated during the inflammatory processes of AIS. However, the microarray meta-analysis in the present study revealed that FYN was significantly downregulated in the blood of patients with AIS. This inconsistent conclusion may be attributed to the dual function of FYN in the inflammatory process, as FYN knockout mice have displayed significantly worse colitis compared with wild-type mice, which correlated with decreased IL-10 and increased IL-17 expression in splenocytes and the gut (52).
Previously, circulating hsa-miR-125a was reported to be upregulated in patients with IS compared with healthy controls (12). A set of circulating miRNAs (hsa-miR-125a-5p, hsa-miR-125b-5p and hsa-miR-143-3p) was demonstrated to have a high accuracy (90.0%), sensitivity (85.6%) and specificity (76.3%) in the diagnosis of patients with IS compared with healthy controls (12). Maitrias et al (53) detected that hsa-miR-125a was significantly overexpressed in stroke patients compared with patients with asymptomatic atherosclerotic carotid plaques. Moreover, Kumar and Nerurkar (54) demonstrated that miR-125a-3p targets genes involved in modulating immune responses, including cyclooxygenase-2, interleukin 1 receptor type 1, IL-10 and C-C motif chemokine ligand 4, in mouse brains (54). Accordingly, it was hypothesized that MCM3AP-AS1 and LINC01089, may be downregulated in AIS, and thus, are unable to sequester hsa-miR-125a and to suppress its inhibitory effects over PXN that contribute to its downregulation. Thus, triggering neuroinflammation in patients' brains and AIS.
The present study has several limitations. Firstly, the sample size of each dataset was small, and additional high-throughput sequencing experiments with larger samples are required to confirm the conclusions. Secondly, the present study only preliminarily screened crucial lncRNA biomarkers for the diagnosis and treatment of AIS. Further clinical experiments, such as quantitative-PCR analysis, correlation analysis, and receiver operating characteristic analysis should be used to prove the diagnostic and prognostic values of the lncRNAs. Finally, the ceRNA mechanisms of DELs should be verified by dual-luciferase reporter assays, and knockout or overexpression in vitro and in vivo.
In conclusion, to the best of our knowledge, the present study is the first to identify several inflammatory related DELs, MCM3AP-AS1, LINC01089, ITPK1-AS1 and HCG27, that may serve as underlying biomarkers for the diagnosis and treatment of AIS. These DELs may function as a ceRNA network (MCM3AP-AS1/LINC01089/hsa-miR-125a/FYN, ITPK1-AS1/hsa-let-7i/RHOA/GRB2/STAT1 and HCG27/hsa-miR-19a/PXN) to induce the development of AIS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The original sequencing data, GSE16561, GSE95204, GSE86291, GSE55937 and GSE110993, were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo). The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LZ, BHL and LH conceived and designed the study. LZ and BHL conducted the statistical analysis. JHH and TTW were involved in the interpretation of the data. LZ and BHL drafted the manuscript. LH participated in critically revising the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Feigin VL, Forouzanfar MH, Krishnamurthi R, Mensah GA, Connor M, Bennett DA, Moran AE, Sacco RL, Anderson L, Truelsen T, et al. Global and regional burden of stroke during 1990–2010: Findings from the Global Burden of disease study 2010. Lancet. 2014;383:3081–255. doi: 10.1016/S0140-6736(13)61953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldsen JK, Engedal TS, Pedraza S, Cho TH, Thomalla G, Nighoghossian N, Baron JC, Fiehler J, Østergaard L, Mouridsen K. Better diffusion segmentation in acute ischemic stroke through automatic tree learning anomaly segmentation. Front Neuroinform. 2018;12:21. doi: 10.3389/fninf.2018.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassella CR, Jagoda A. Ischemic Stroke: Advances in diagnosis and management. Emerg Med Clin North Am. 2017;35:911–930. doi: 10.1016/j.emc.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Hasan TF, Rabinstein AA, Middlebrooks EH, Haranhalli N, Silliman SL, Meschia JF, Tawk RG. Diagnosis and management of acute ischemic stroke. Mayo Clin Proc. 2018;93:523–538. doi: 10.1016/j.mayocp.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127:761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu M, Li N, Luo P, Jing W, Wen X, Liang C, Tu J. Peripheral blood leukocyte expression of lncRNA MIAT and its diagnostic and prognostic value in ischemic stroke. J Stroke Cerebrovasc Dis. 2017;27:326–337. doi: 10.1016/j.jstrokecerebrovasdis.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Feng L, Guo J, Ai F. Circulating long noncoding RNA ANRIL downregulation correlates with increased risk, higher disease severity and elevated pro-inflammatory cytokines in patients with acute ischemic stroke. J Clin Lab Anal. 2019;33:e22629. doi: 10.1002/jcla.22629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng QW, Li S, Wang H, Sun HL, Zuo L, Gu ZT, Lu G, Sun CZ, Zhang HQ, Yan FL. Differential long noncoding RNA expressions in peripheral blood mononuclear cells for detection of acute ischemic stroke. Clin Sci. 2018;132:1597–1614. doi: 10.1042/CS20180411. [DOI] [PubMed] [Google Scholar]
- 9.Sheng Y, Ma J, Zhao J, Qi S, Hu R, Yang Q. Differential expression patterns of specific long noncoding RNAs and competing endogenous RNA network in alopecia areata. J Cell Biochem. 2019;120:10737–10747. doi: 10.1002/jcb.28365. [DOI] [PubMed] [Google Scholar]
- 10.Guo X, Yang J, Liang B, Shen T, Yan Y, Huang S, Zhou J, Huang J, Gu L, Su L. Identification of novel LncRNA biomarkers and construction of LncRNA-related networks in Han Chinese patients with ischemic stroke. Cell Physiol Biochem. 2018;50:2157–2175. doi: 10.1159/000495058. [DOI] [PubMed] [Google Scholar]
- 11.He W, Wei D, Chen S, Li S, Chen W. Altered long non-coding RNA transcriptomic profiles in ischemic stroke. Hum Gene Ther. 2017;29:719–732. doi: 10.1089/hum.2017.064. [DOI] [PubMed] [Google Scholar]
- 12.Tiedt S, Prestel M, Malik R, Schieferdecker N, Duering M, Kautzky V, Stoycheva I, Böck J, Northoff BH, Klein M, et al. RNA-Seq identifies circulating miR-125a-5p, miR-125b-5p and miR-143-3p as potential biomarkers for acute ischemic stroke. Circ Res. 2017;121:970–980. doi: 10.1161/CIRCRESAHA.117.311572. [DOI] [PubMed] [Google Scholar]
- 13.Tian C, Li Z, Yang Z, Huang Q, Liu J, Hong B. Plasma MicroRNA-16 is a biomarker for diagnosis, stratification, and prognosis of hyperacute cerebral infarction. PLoS One. 2016;11:e0166688. doi: 10.1371/journal.pone.0166688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jickling GC, Ander BP, Xinhua Z, Dylan N, Boryana S, Dazhi L. microRNA expression in peripheral blood cells following acute ischemic stroke and their predicted gene targets. PLoS One. 2014;9:e99283. doi: 10.1371/journal.pone.0099283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Yanqing Z, Yaru C, Jia G. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43((Database Issue)):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashwini J, Marks DS, Erik L. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42((Database Issue)):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM, Hatzigeorgiou AG. DIANA-LncBase: Experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41((Database Issue)):D239–D245. doi: 10.1093/nar/gks1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dweep H, Gretz N. miRWalk2. 0: A comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 25.Jin R, Liu L, Zhang S, Nanda A, Li G. Role of inflammation and its mediators in acute ischemic stroke. J Cardiovasc Transl Res. 2013;6:834–851. doi: 10.1007/s12265-013-9508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dziedzic T. Systemic inflammation as a therapeutic target in acute ischemic stroke. Expert Rev Neurother. 2015;15:523–531. doi: 10.1586/14737175.2015.1035712. [DOI] [PubMed] [Google Scholar]
- 27.Akella A, Bhattarai S, Dharap A. Long noncoding RNAs in the pathophysiology of ischemic stroke. Neuromolecular Med. 2019;21:474–483. doi: 10.1007/s12017-019-08542-w. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Zhu XL, Ji BY, Cao X, Yu LJ, Zhang Y, Bao XY, Xu Y, Jin JL. LncRNA-1810034E14Rik reduces microglia activation in experimental ischemic stroke. J Neuroinflammation. 2019;16:75. doi: 10.1186/s12974-019-1464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Zhao H, Fan Z, Li G, Ma Q, Tao Z, Wang R, Feng J, Luo Y. Long noncoding RNA H19 promotes neuroinflammation in ischemic stroke by driving histone deacetylase 1-Dependent M1 microglial polarization. Stroke. 2017;48:2211–2221. doi: 10.1161/STROKEAHA.117.017387. [DOI] [PubMed] [Google Scholar]
- 30.Cao DW, Liu MM, Duan R, Tao YF, Zhou JS, Fang WR, Zhu JR, Niu L, Sun JG. The lncRNA Malat1 functions as a ceRNA to contribute to berberine-mediated inhibition of HMGB1 by sponging miR-181c-5p in poststroke inflammation. Acta Pharmacol Sin. 2020;41:22–33. doi: 10.1038/s41401-019-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Luo Y, Yao Y, Ji Y, Feng L, Du F, Zheng X, Tao T, Zhai X, Li Y, et al. Silencing the lncRNA Maclpil in pro-inflammatory macrophages attenuates acute experimental ischemic stroke via LCP1 in mice. J Cereb Blood Flow Metab. 2020;40:747–759. doi: 10.1177/0271678X19836118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K, Qi M, Yang Y, Xu P, Zhua Y, Zhang J. Circulating lncRNA ANRIL in the serum of patients with ischemic stroke. Clin Lab. 2019:65. doi: 10.7754/Clin.Lab.2019.190143. doi: 10.7754/Clin.Lab.2019.190143. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu Q, Tong X, Yang W, Xu Q, Huang D, Tu K. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer Res. 2019;18:28. doi: 10.1186/s12943-019-0957-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sas-Chen A, Aure MR, Leibovich L, Carvalho S, Enuka Y, Körner C, Polycarpou-Schwarz M, Lavi S, Nevo N, Kuznetsov Y, et al. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO Mol Med. 2016;8:1052–1064. doi: 10.15252/emmm.201606198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Z, Yang D, Tang Y, Zhang X, Wei Z, Fu H, Xu J, Zhu Z, Cai Q. Five-long non-coding RNA risk score system for the effective prediction of gastric cancer patient survival. Oncol Lett. 2019;17:4474–4486. doi: 10.3892/ol.2019.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang C, Zheng J, Xue Y, Yu H, Liu X, Ma J, Liu L, Wang P, Li Z, Cai H, Liu Y. The Effect of MCM3AP-AS1/miR-211/KLF5/AGGF1 axis regulating glioblastoma angiogenesis. Front Mol Neurosci. 2017;10:437. doi: 10.3389/fnmol.2017.00437. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Ding H, Gao S, Wang L, Wei Y, Zhang M. Overexpression of miR-582-5p inhibits the apoptosis of neuronal cells after cerebral ischemic stroke through regulating PAR-1/Rho/Rho Axis. J Stroke Cerebrovasc Dis. 2019;28:149–155. doi: 10.1016/j.jstrokecerebrovasdis.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 38.Vesterinen HM, Currie GL, Carter S, Mee S, Watzlawick R, Egan KJ, Macleod MR, Sena ES. Systematic review and stratified meta-analysis of the efficacy of RhoA and Rho kinase inhibitors in animal models of ischaemic stroke. Syst Rev. 2013;2:33. doi: 10.1186/2046-4053-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schultz NEØ, Hasseldam H, Rasmussen RS, Vindegaard N, Mcwilliam O, Iversen HK, Johansen FF. Statin treatment before stroke reduces pro-inflammatory cytokine levels after stroke. Neurol Res. 2019;41:289–297. doi: 10.1080/01616412.2018.1558000. [DOI] [PubMed] [Google Scholar]
- 40.Fard MA, Ebrahimi KB, Miller NR. RhoA activity and post-ischemic inflammation in an experimental model of adult rodent anterior ischemic optic neuropathy. Brain Res. 2013;1534:76–86. doi: 10.1016/j.brainres.2013.07.053. [DOI] [PubMed] [Google Scholar]
- 41.Jin K, Mao X, Mw, Nagayama T, Minami M, Simon R, Greenberg D. Microarray analysis of hippocampal gene expression in global cerebral ischemia. Ann Neurol. 2010;50:93–103. doi: 10.1002/ana.1073. [DOI] [PubMed] [Google Scholar]
- 42.He D, Zhuo Z, Lao J, Meng H, Han L, Fan C, Dan Y, He Z, Yun X. Proteomic analysis of the Peri-infarct area after human umbilical cord mesenchymal stem cell transplantation in experimental stroke. Aging Dis. 2016;7:623–634. doi: 10.14336/AD.2016.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou YC, Liou KT, Chern CM, Wang YH, Liao JF, Chang S, Chou YH, Shen YC. Preventive effect of silymarin in cerebral ischemia-reperfusion-induced brain injury in rats possibly through impairing NF-κB and STAT-1 activation. Phytomedicine. 2010;17:963–973. doi: 10.1016/j.phymed.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 44.Xiang W, Tian C, Lin J, Wu X, Pang G, Zhou L, Pan S, Deng Z. Plasma let-7i and miR-15a expression are associated with the effect of recombinant tissue plasminogen activator treatment in acute ischemic stroke patients. Thromb Res. 2017;158:121–125. doi: 10.1016/j.thromres.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 45.Jickling GC, Ander BP, Shroff N, Orantia M, Stamova B, Dykstra-Aiello C, Hull H, Zhan X, Liu D, Sharp FR. Leukocyte response is regulated by microRNA let7i in patients with acute ischemic stroke. Neurology. 2016;87:2198–2205. doi: 10.1212/WNL.0000000000003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franciska E, Thorsten T, Günter M, Konstantin-A H. Immunohistochemical analysis of protein expression after middle cerebral artery occlusion in mice. Acta Neuropathol. 2004;107:127–136. doi: 10.1007/s00401-003-0789-8. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Zhou CX, Zhang ZJ, Wang LY, Jing ZW, Wang Z. Synergistic mechanism of gene expression and pathways between jasminoidin and ursodeoxycholic acid in treating focal cerebral ischemia-reperfusion injury. CNS Neurosci Ther. 2012;18:674–682. doi: 10.1111/j.1755-5949.2012.00348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan JR, Tan KS, Yong FL, Armugam A, Wang CW, Jeyaseelan K, Wong PT. MicroRNAs regulating cluster of differentiation 46 (CD46) in cardioembolic and non-cardioembolic stroke. PLoS One. 2017;12:e0172131. doi: 10.1371/journal.pone.0172131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marie JC, Astier AL, Rivailler P, Rabourdin-Combe C, Wild TF, Horvat B. Linking innate and acquired immunity: Divergent role of CD46 cytoplasmic domains in T cell induced inflammation. Nat Immunol. 2002;3:659–666. doi: 10.1038/ni810. [DOI] [PubMed] [Google Scholar]
- 50.Du CP, Tan R, Hou XY. Fyn kinases play a critical role in neuronal apoptosis induced by oxygen and glucose deprivation or amyloid-β peptide treatment. CNS Neurosci Ther. 2012;18:754–761. doi: 10.1111/j.1755-5949.2012.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holmes A, Zhou N, Donahue DL, Balsara R, Castellino FJ. A deficiency of the GluN2C subunit of the N-methyl-D-aspartate receptor is neuroprotective in a mouse model of ischemic stroke. Biochem Biophys Res Commun. 2018;495:136–144. doi: 10.1016/j.bbrc.2017.10.171. [DOI] [PubMed] [Google Scholar]
- 52.Lopes F, Wang A, Smyth D, Reyes JL, Doering A, Schenck LP, Beck P, Waterhouse C, McKay DM. The Src kinase Fyn is protective in acute chemical-induced colitis and promotes recovery from disease. J Leukoc Biol. 2015;97:1089–1099. doi: 10.1189/jlb.3A0814-405RR. [DOI] [PubMed] [Google Scholar]
- 53.Maitrias P, Metzinger-Le Meuth V, Massy ZA, M'Baya-Moutoula E, Reix T, Caus T, Metzinger L. MicroRNA deregulation in symptomatic carotid plaque. J Vasc Surg. 2015;62:1245–1250.e1. doi: 10.1016/j.jvs.2015.06.136. [DOI] [PubMed] [Google Scholar]
- 54.Kumar M, Nerurkar VR. Integrated analysis of microRNAs and their disease related targets in the brain of mice infected with West Nile virus. Virology. 2014;452-453:143–151. doi: 10.1016/j.virol.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original sequencing data, GSE16561, GSE95204, GSE86291, GSE55937 and GSE110993, were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo). The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.