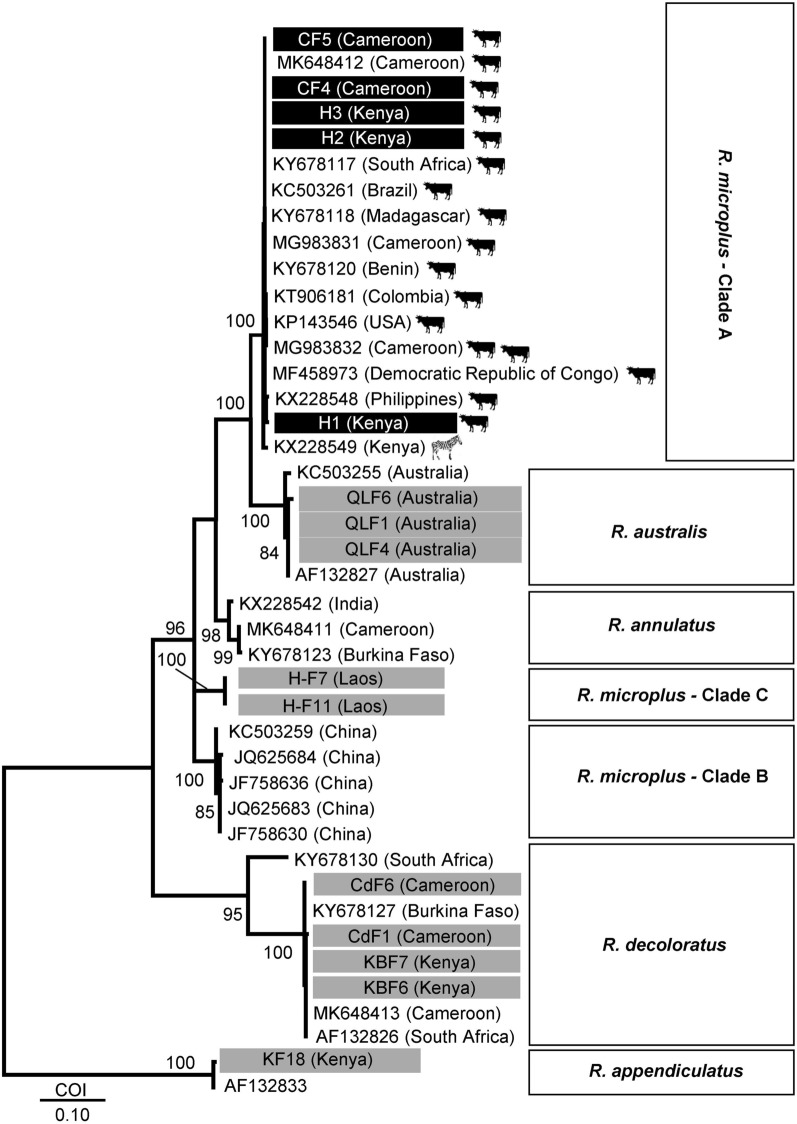
Fig. 6.
Maximum Likelihood (ML) cox1 tree showing the phylogenetic relationships between ticks analysed in this study and 28 reference sequences. The tree was constructed based on Tamura 3-parameter (T92) model [51]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 69.54% sites). This analysis involved 42 nucleotide sequences. Tick samples analysed in this study are highlighted. The scale represents 0.10 nucleotide substitutions per site. There was a total of 403 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [31]. Bootstrap values (1000 replications) above 70% are shown