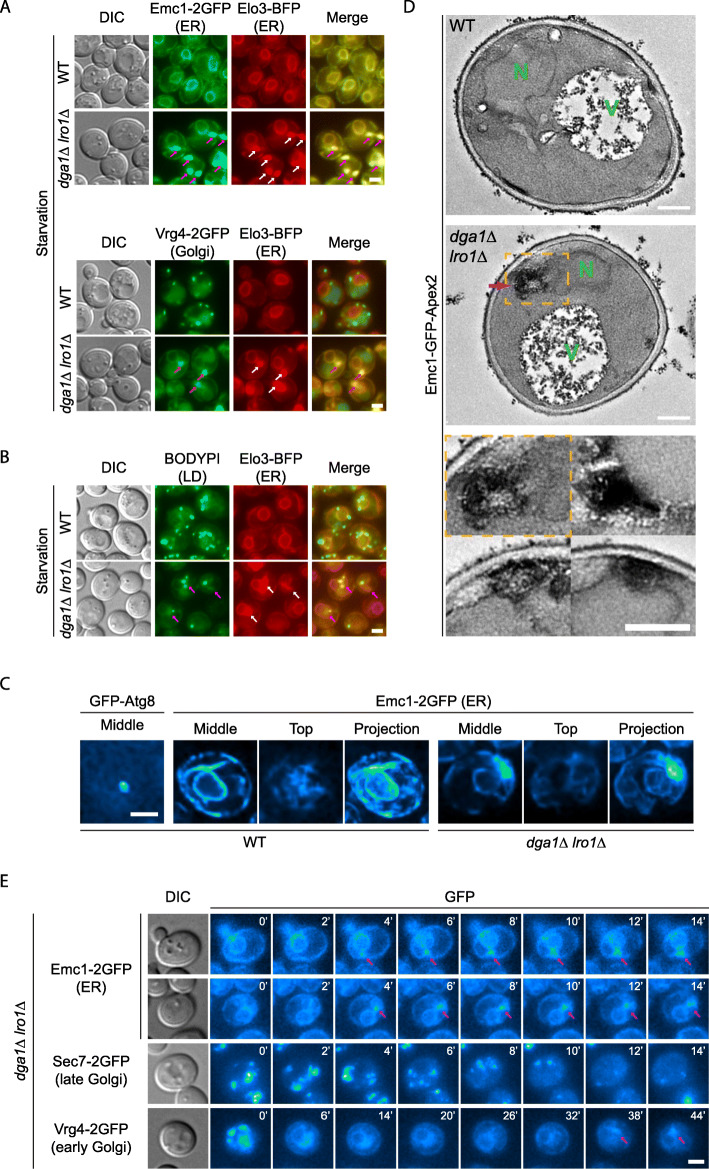
Fig. 2.
Characterization of endomembrane system defects in TAG production defective cells. a The bulbous ER structure contains multiple ER and Golgi proteins. Cells co-expressing two protein chimeras as indicated were starved for 1 h. Representative image slices from individual channels and merged channels are shown. Arrows, bulbous structures on the ER. Scale bar, 2 μm. b The bulbous ER structures are not lipid droplets. Cells expressing Elo3-BFP were starved for 1 h and stained with BODIPY. Images presented as in a. c The ER bulbs are not hollow vesicles. Cells were starved for 1 h. Autophagosomes and ER bulbs were imaged using the Super Resolution via Optical Re-assignment (SORA) technique. Scale bar, 2 μm. d Electron micrograph of yeast cells carrying the indicated genotypes. Cells expressing Emc1-GFP-Apex2 were starved for 1 h and stained with DAB. Representative transmission electron micrographs from two independent repeats are shown. N, nucleus. V, vacuoles. Purple arrows, electron-dense structures connected to the ER. Inserts below, magnified view of demarcated area above and three additional areas from dga1Δ lro1Δ samples, showing the electron-dense structures. Scale bar, 0.5 μm. e Time-lapse imaging of the endomembrane defects. dga1Δ lro1Δ cells were transferred from rich medium to nitrogen starvation medium. Representative image slices (Emc1) or projections (Sec7, Vrg4) at the indicated time point are shown. Time 0 corresponds to 20 min after the medium transfer. Scale bar, 2 μm