Abstract
The mTOR pathway serves an important role in the development of insulin resistance induced by obesity. Exercise improves obesity-associated insulin resistance and hepatic energy metabolism; however, the precise mechanism of this process remains unknown. Therefore, the present study investigated the role of rapamycin, an inhibitor of mTOR, on exercise-induced expression of hepatic energy metabolism genes in rats fed a high-fat diet (HFD). A total of 30 male rats were divided into the following groups: Normal group (n=6) fed chow diets and HFD group (n=24) fed an HFD for 6 weeks. The HFD rats performed exercise adaptation for 1 week and were randomly divided into the four following groups (each containing six rats): i) Group of HFD rats with sedentary (H group); ii) group of HFD rats with exercise (HE group); iii) group of HFD rats with rapamycin (HR group); and iv) group of HFD rats with exercise and rapamycin (HER group). Both HE and HER rats were placed on incremental treadmill training for 4 weeks (from week 8–11). Both HR and HER rats were injected with rapamycin intraperitoneally at the dose of 2 mg/kg once a day for 2 weeks (from week 10–11). All rats were sacrificed following a 12–16 h fasting period at the end of week 11. The levels of mitochondrial and oxidative enzyme activities, as well as of the expression of genes involved in energy metabolism were assessed in liver tissues. Biochemical assays and oil red staining were used to assess the content of hepatic triglycerides (TGs). The results indicated that exercise, but not rapamycin, reduced TG content in the liver of HFD rats. Further analysis indicated that rapamycin reduced the activity of cytochrome c oxidase, but not the activities of succinate dehydrogenase and β-hydroxyacyl-CoA dehydrogenase in the liver of HFD rats. Exercise significantly upregulated the mRNA expression of peroxisome proliferator-activated receptor γ coactivator 1 β, while rapamycin exhibited no effect on the mRNA expression levels of hepatic transcription factors associated with energy metabolism enzymes in the liver of HFD rats. Collectively, the results indicated that exercise reduced TG content and upregulated mitochondrial metabolic gene expression in the liver of HFD rats. Moreover, this mechanism may not involve the mTOR pathway.
Keywords: rapamycin, mTOR, obesity, exercise, mitochondrial oxidative metabolism
Introduction
A chronic excessive intake of fat is associated with its accumulation in various tissues. For instance, this process does not only occur in adipose tissues, but also in the liver, skeletal muscle and other non-adipose tissues, leading to the development of obesity and insulin resistance (1,2). Exercise improves insulin resistance by reducing obesity-induced fat accumulation in adipose tissues and ectopic deposition of fat in the liver (3,4). Moreover, it has been reported that mTOR serves an important role in the development of insulin resistance induced by obesity (5). mTOR is a highly conserved serine/threonine kinase, which includes two functionally distinct multiprotein complexes, known as the rapamycin-sensitive mTOR complex 1 (mTORC1) and rapamycin-insensitive mTOR complex 2 (mTORC2) (5). mTORC1 leads to feedback suppression of the insulin signals and of its substrate ribosomal protein S6 kinase 1 (S6K1). Furthermore, high-fat diet (HFD)-induced obesity constitutively activates mTORC1 signaling pathways in the liver, leading to cellular damage, while exercise markedly lowers the increased activities of mTORC1 signaling in the liver caused by HFD (6–8). The mTORC1 inhibitor rapamycin, which is used as a potent immunosuppressant in patients with organ transplants, has been considered for the treatment of metabolic disorders (9). However, the precise mechanisms underlying the improvement of insulin resistance induced by exercise and the maintenance of hepatic lipid homeostasis in HFD remain unknown.
mTORC1 is an important sensor that allows cells and tissues to adapt to their metabolism in response to nutritional stimuli (10). It has been shown that nuclear transcription factors, including peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1), peroxisome proliferator-activated receptors (PPARs), sterol regulatory element binding protein-1c (SREBP-1c) and carbohydrate-response element-binding protein (ChREBP), are critical to the regulation of liver glucose and lipid metabolism gene expression, which in turn affects the activity of the enzymes involved in fatty acid metabolism (11,12). mTORC1 has been identified as a positive regulator of hepatic triglyceride synthesis and a negative regulator of the expression levels of hepatic PGC-1α, PPARα and its target genes, such as carnitine palmitoyl transferase-1 (CPT1) and pyruvate dehydrogenase kinase 4 (PDK4) (13,14). Exercise can promote glucose and lipid homeostasis, as well as improve glucose tolerance and insulin intolerance (15). Chronic administration of rapamycin may led to dysregulated lipid and glucose metabolism, causing a reduction of fat mass and the aggravation of glucose intolerance (16). Previous studies have investigated the changes noted in hepatic lipid and glucose metabolism in response to mTOR activation or inactivation based on liver-specific knockout models of phosphatase and tensin homolog, Tuberous Sclerosis Complex-1 (Tsc1), mTORC1 component and rapamycin administration in different contexts (17–19). For example, the liver-specific deletion of DEP domain containing mTOR-interacting protein increased the activity of mTORC1, thereby promoting oxidative metabolism and reducing circulating glucose levels in fasting mice, which were corrected by rapamycin (17). In addition, the activation of mTORC1 induced by the loss of hepatic Tsc1, suppressed gluconeogenesis and tricarboxylic acid cycle oxidation (18).
In the present study, biochemical, molecular biological and histological methods were used to investigate the effects of rapamycin on the exercise-induced changes caused in genes associated with energy metabolism in the liver of HFD rats.
Materials and methods
Experimental animals
A total of 30 male Sprague-Dawley rats (weight, 150–180 g; age, 5–6 weeks) were purchased from Guangdong Province Experimental Animal Center. All the experimental procedures were approved by the Institutional Animal Care and Use Committee of the Guangzhou Sport University. All rats were raised in specific pathogen free animal laboratory of Guangzhou Institute of Sports Science. Rats were housed under standardized conditions with stable temperature (22–25°C) and humidity (60±5%), a controlled light/dark cycle (light: 06:00-18:00; dark: 18:00-06:00) and free access to water and food. After 1 week of adaptation maintenance, rats were randomized into normal group (N; n=6) fed chow diet and HFD group (n=24) fed HFD (20% carbohydrate, 59% fat, 21% protein) (20) for 6 weeks.
At week 7, HFD rats were given an adaptive exercise training by running on a motor-driven rodent treadmill (tail electro-stimulation) for 15 min per day for 4 days at a speed of 15 m/min (0° incline), then randomly subdivided into the following groups (n=6 rats/group): i) HFD rats with sedentary (H group); ii) HFD rats with exercise (HE group); iii) HFD rats with rapamycin (HR group); and iv) HFD rats with exercise and rapamycin (HER group).
Exercise program and treatment
From weeks 8–11, the rats in the HE and HER groups were running on a treadmill for 20–40 min once a day, 5 days per week. The exercise volume consisted of 20–23 m/min (20–30 min) at 0% slope on week 1, 23–25 m/min (30–40 min) at 0% slope on week 2 and 25 m/min (40 min) at 0% slope on weeks 3–4. From the 10th week, the rats in the HR and HER groups received intraperitoneal (IP) injection of rapamycin (2 mg/kg daily; Sigma-Aldrich; Merck KGaA) at an exercising day (1 h prior exercise) for 2 weeks (5 days/week). In the last 2 days of the 11th week, the Glucose Tolerance Test (GTT) and the Insulin Tolerance Test (ITT) were performed. GTT was performed by IP injection of glucose (2 g/kg body weight; Sigma-Aldrich; Merck KGaA) following fasting for 16 h. Blood glucose was measured at 0, 30, 60 and 120 min in 10–20 µl blood obtained via tail nicking using the GLUCO™ CardII glucometer (GT-1640). Similarly, ITT was performed via IP injection of regular insulin (0.75 U/kg; Novo Nordisk) followed by glucose measurements at 0, 15, 30 and 60 min.
At the end of the 11th week, all animals were fasted for 12–16 h and subsequently anesthetized with pentobarbital sodium (50 mg/kg) via IP injection. After rats were deeply anesthetized, 4–6 ml arterial blood from the abdominal aorta was collected and the plasma was immediately separated using centrifugation (1,000 × g for 10 min at room temperature) and stored at −80°C for biochemical analysis. Then, rats were euthanatized via cervical dislocation. The liver tissues were quickly excised and fixed with 4% paraformaldehyde at room temperature for 48 h or frozen in liquid nitrogen and subsequently stored at −80°C for further analysis. Visceral fat weight was removed and estimated by the weight of greater mesenteric, epididymal and perirenal fat (20). The experimental procedure is presented in Fig. 1. All animals maintained good health throughout the experiment, with no need for early euthanasia.
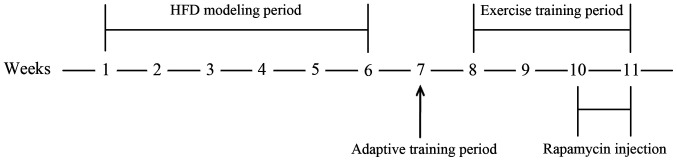
Figure 1.
A schematic diagram of the experimental procedure. The 1st to 6th week was the high-fat diet modeling period, the 7th week was the adaptive training period, the 8th to the 11th week was the exercise training period and the 10th to 11th week was the rapamycin injection period. HFD, high-fed diet.
Blood biochemical analysis
Fasting blood glucose (FBG; cat. no. F006-1-1), fasting plasma TG (FTG; cat. no. A110-2-1) and fasting plasma non-esterified fatty acids (FFA; cat. no. A042-1-1) were detected using colorimetric assay kits (Nanjing Jiancheng Bioengineering Institute). Fasting plasma insulin (FINS; cat. no. MM-0587R1) and β-hydroxybutyrate (cat. no. MM-20814R1) were measured according to the manufacturers' protocols of commercially available ELISA kits (Jiangsu Meimian industrial Co., Ltd.).
Measurement of hepatic TG content and mitochondrial enzyme activities
The TG content in the liver was determined following lipid extraction using the Folch method (21). The mitochondrial enzyme activities were measured as follows: A portion of liver tissue was homogenized in ice-cold buffer containing 175 mM KCl, 10 mM glutathione and 2 mM EDTA at pH 7.4. The homogenized extracts were centrifuged at 18,000 × g for 5 min at 4°C and the supernatants were transferred to new tubes. Succinate dehydrogenase (SDH; cat. no. K660-100) and cytochrome c oxidase (COX; cat. no. K287-100) activities were measured at 37°C using commercially available kits according to the manufacturers' instructions (BioVision, Inc.). β-hydroxyacyl-CoA dehydrogenase (HADH) activity was determined at 37°C using the reaction media containing 0.1 M triethanolamine-HCl, 5 mM EDTA, 0.45 mM NADH and 0.1 mM acetoacetyl-CoA at pH 7.0 (22). Enzyme activity was assayed spectrophotometrically using the Tecan Microplate Reader (Infinite M200; Tecan Group, Ltd.).
Hematoxylin and eosin (H&E) and Oil Red O staining
For H&E staining, the liver tissue was fixed with 4% paraformaldehyde for 48 h at room temperature, dehydrated with an ascending alcohol series, made transparent in xylene at room temperature and embedded in paraffin. Subsequently, the samples were cut into 12-µm thick cross sections using a rotary microtome (Leica Microsystems GmbH). The sections were washed with xylene at room temperature, rehydrated using a descending alcohol series and stained with hematoxylin (Sigma-Aldrich; Merck KGaA) for 15 min at room temperature. Subsequently, the sections were washed with water for 10 min, immersed in 0.2% hydrochloric acid alcohol for 3–5 sec, washed with water for another 10 min and then stained with 1% eosin (Sigma-Aldrich; Merck KGaA) for 1 min at room temperature. Following the staining, the sections were dehydrated with an ascending alcohol series, washed with xylene and mounted with neutral balsam (Beijing Solarbio Science & Technology Co., Ltd.).
For Oil Red O staining, the liver tissue was fixed in 4% paraformaldehyde for 48 h at room temperature, rinsed with 0.1 M PBS for 10 min at room temperature, dehydrated with an ascending sucrose series at 4°C, embedded in OCT-freeze medium (cat. no. 4583; Sakura Finetek USA, Inc.) and sectioned in 12-µm thick sections using a freezing microtome (Leica Microsystems GmbH). Subsequently, the sections were washed with deionized water 3 times for 30 sec each, stained with a working solution of 0.3% Oil red O (cat. no. O0625; Sigma-Aldrich; Merck KGaA) at room temperature for 30 min, rinsed with water for 10 min and then mounted with glycerol gelatin (Sigma-Aldrich; Merck KGaA). Stained sections were visualized using a light microscope (magnification, ×100 and ×200; LEICA DM750; Leica Microsystems GmbH).
Reverse transcription-quantitative PCR (RT-qPCR)
The mRNA expression levels of PPARα, PPARβ, PGC-1α, PGC-1β, SREBP-1c, ChREBP, CPT1α, acyl-CoA carboxylase 1(ACCα) and PDK4 were detected in the liver tissues using RT-qPCR (LightCycler®; Roche Diagnostic). Total RNA was extracted from 50 mg liver tissue using an RNA extraction reagent (cat. no. CW0580; CoWin Biosciences). Following spectroscopic quantification, cDNA was synthesized using 4 µl RNA template at the following conditions: 37°C for 15 min and 85°C for 5 sec using the PrimeScript™ RT reagent kit (cat. no. RR047A; Takara Bio, Inc.). Subsequently, 9.5 µl dH2O, 12.5 µl TB Green Premix Ex Taq buffer (2X), 0.5 µl forward primer (10 pmol/µl), 0.5 µl reverse primer (10 pmol/µl) and 2 µl cDNA were mixed based on the manufacturer's protocol of the TB Green Premix Ex Taq™ kit (cat. no. RR420A; Takara Bio, Inc.), and RT-qPCR was performed using the following conditions: Initial denaturation at 93°C for 2 min, followed by 40 cycles at 93°C for 15 sec, 55°C for 25 sec and 72°C for 25 sec. The primer sequences of each gene are listed in Table SI. The Cycle Threshold values, which represented the cycle times when the fluorescence signal in each reaction tube reached the set threshold value, were calculated. The housekeeping gene GAPDH was used in order to normalize the expression levels for each gene of interest according to the mean cycle number. The 2−ΔΔCq method was used to calculate the relative amounts of the target gene (23).
Western blotting
The liver tissues were lysed with a solution containing 150 mM NaCl, 5 mM EDTA, 50 mM Tri-HCl (pH 8.0), 1% NP 40, 1 mM aprotinin, 0.1 mM leupeptin and 1 mM pepstatin. The solution was centrifuged at 4°C at 18,000 × g for 30 min. The supernatants were collected and the protein concentration in the homogenates was measured using the Bradford method (24). Subsequently, 30 µg protein/lane was separated using 10% SDS-PAGE, transferred to nitrocellulose membranes, blocked in 5% skimmed milk for 1 h at room temperature, incubated with the primary antibodies overnight at 4°C and then incubated with secondary antibodies for 2 h at room temperature. The primary antibodies used were as follows: Anti-akt (1:200; cat. no. sc-5298; Santa Cruz Biotechnology, Inc.), anti-phosphorylated (p)-Akt ser473(1:1,000; cat. no. AF887; Bio-Techne Ltd.), anti-S6 (1:200; cat. no. sc-8418; Santa Cruz Biotechnology, Inc.), anti-p-S6 (1:200; cat. no. sc-54279; Santa Cruz Biotechnology, Inc.) and anti-GAPDH (1:3,000; cat. no. CW0100M; CoWin Biosciences, Inc.). The fluorescent secondary antibodies were as follows: IRDye 680LT-conjugated anti-rabbit (red; 1:20,000; cat. no. 925-68021; LI-COR Biosciences) and IRDye 800-conjugated anti-mouse secondary antibody (green; 1:20,000; cat. no. 925-32212; LI-COR Biosciences). The membrane bands were scanned and assessed using the LI-COR Odyssey infrared imaging system (LI-COR Biosciences). The expression levels were analyzed using LI-COR Odyssey application software (version 3.0.30; LI-COR Biosciences).
Statistical analysis
One-way ANOVA followed by Bonferroni post hoc test was performed for any intergroup differences. All of the analyses were performed using the SPSS 21.0 software (IBM Corp.). Data are presented as the mean ± standard error. P<0.05 was considered to indicate a statistically significant difference.
Results
Exercise but not rapamycin reduces TG content in the liver of HFD rats
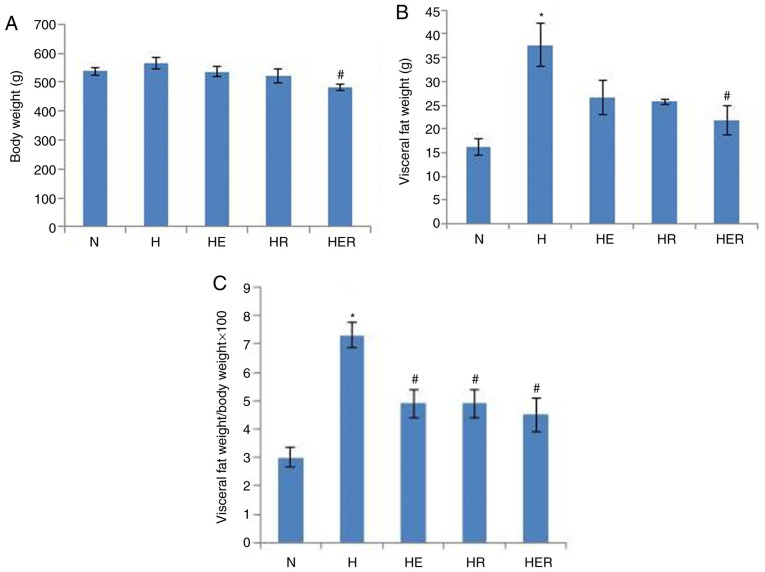
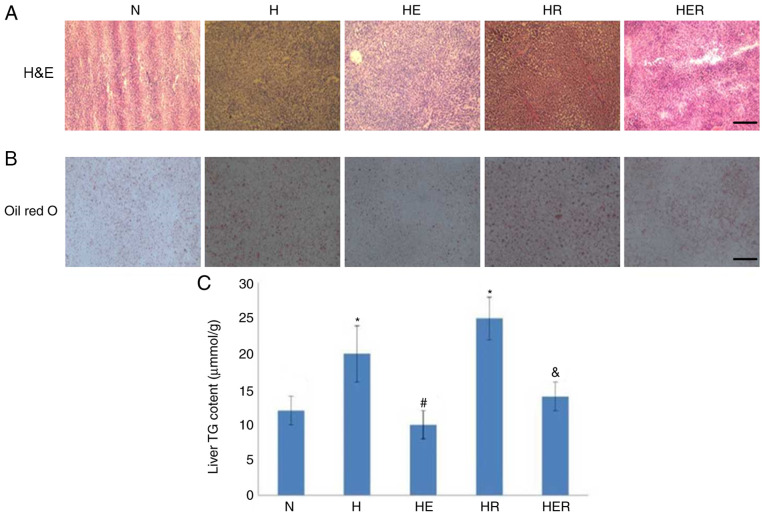
Compared with the N group, the visceral fat weight was significantly increased in the H group (Fig. 2B). HER rats exhibited significantly lower body weight and visceral fat compared with the H rats (Fig. 2A-C). However, exercise and rapamycin exhibited no significant effects on the levels of blood biochemical parameters (Table SII). Histological analysis and biochemical measurements indicated significant lipid deposition in the rats of the H group compared with rats in the N group. In contrast, compared with the H group, lipid deposition was markedly reduced in the HE group. Compared with the HR group, lipid deposition was markedly reduced in the HER group. These results indicated that exercise can reduce lipid deposition (Fig. 3).
Figure 2.
Body weight and visceral fat changes. (A) Body weight of each group. (B) Visceral fat weight of each group. (C) Ratio of visceral fat weight to body weight in each group. Data are presented as the mean ± SE for n=4–6 rats. *P<0.05 vs. N group; #P<0.05 vs. H group. N, normal; H, HFD rats with sedentary; HFD, high-fed diet; E, exercise; R, rapamycin.
Figure 3.
Histological tests and TG content test in the liver. (A) H&E staining. Scale bar, 100-µm. (B) Oil red O staining. Scale bar, 200-µm. (C) Hepatic TG content test. Data are presented as the mean ± SE for n=4-6 rats. *P<0.05 vs. N group; #P<0.05 vs. H group; &P<0.05 vs. HR group. H&E, hematoxylin and eosin; N, normal; H, HFD rats with sedentary; HR, HFD rats with rapamycin; HFD, high-fed diet; E, exercise; TG, triglyceride.
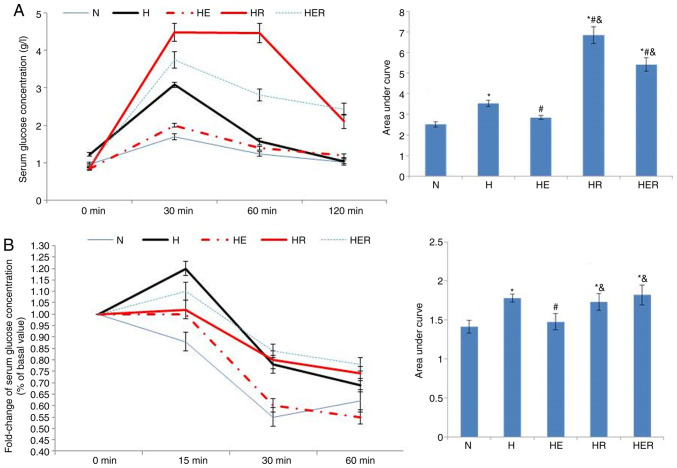
In the glucose tolerance test, the serum glucose concentration was significantly increased in the H, HR and HER groups compared with the N group (Fig. 4A). Compared with the H group, the serum glucose concentration was markedly reduced in the HE group, while increased in the HR and HER groups. Compared with the HE group, the serum glucose concentration was increased in the HR and HER groups. In the insulin tolerance test, the fold-change of serum glucose concentration was significantly increased in the H, HR and HER groups compared with the N group (Fig. 4B). Compared with the H group, the fold change of serum glucose concentrations in the HE group was significantly reduced. Compared with the HE group, the fold change of serum glucose concentrations in the HR and HER groups were significantly increased. These results indicated that exercise significantly alleviated glucose intolerance and insulin intolerance, while rapamycin not only increased glucose intolerance, but also impaired exercise-induced improvement of glucose intolerance and insulin intolerance (Fig. 4).
Figure 4.
Glucose tolerance and insulin tolerance tests. (A) Glucose tolerance test. (B) Insulin tolerance tests. Data are presented as the mean ± SE for n=4 rats. *P<0.05 vs. N group; #P<0.05 vs. H group; &P<0.05 vs. HE group. N, normal; H, HFD rats with sedentary; HE, HFD rats with exercise; HFD, high-fed diet; E, exercise; R, rapamycin.
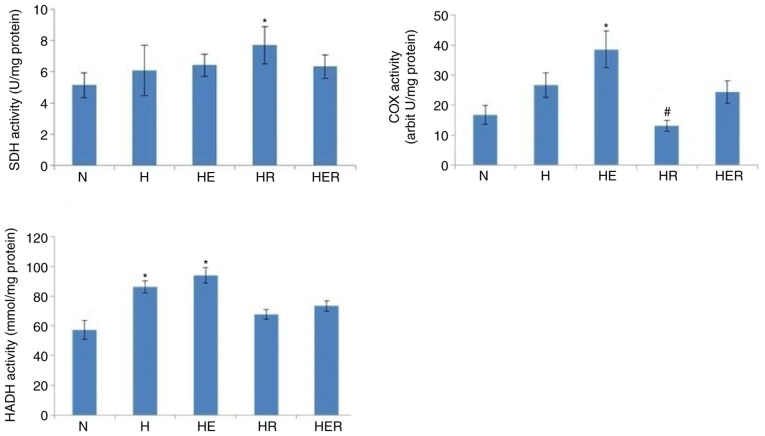
Effects of exercise and rapamycin on mitochondrial enzyme activities
Exercise increased the activities of COX and HADH, but not those of SDH, in the liver of HFD rats compared with the N group. In addition, compared with the H group, rapamycin significantly reduced COX activity, while it exhibited no effects on the activities of SDH and HADH in the liver tissues of HFD rats (Fig. 5).
Figure 5.
Hepatic mitochondrial enzyme activities in rats fed N chow or HFD, with or without E following R administration. Data are presented as the mean ± SE for n=4-6 rats. *P<0.05 vs. N group. #P<0.05 vs. H group. N, normal; H, HFD rats with sedentary; HFD, high-fed diet; E, exercise; R, rapamycin; SDH, Succinate dehydrogenase; COX, cytochrome c oxidase; HADH, β-Hydroxyacyl-CoA dehydrogenase.
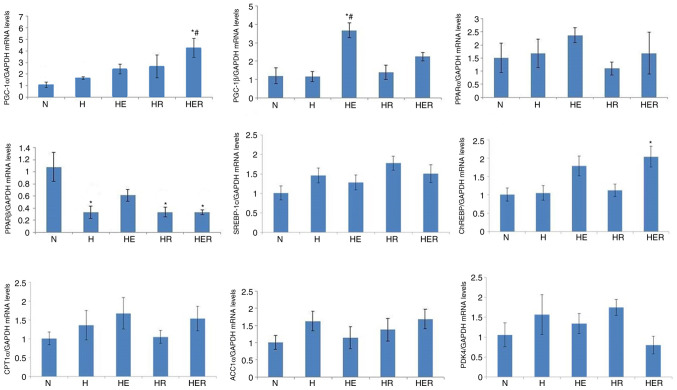
Effects of exercise and rapamycin on the expression levels of energy metabolism genes in the liver
The mRNA expression of PGC-1α in the HER group was significantly increased compared with the N and H groups. Moreover, hepatic PPARβ mRNA expression was significantly reduced in the H, HR, and HER groups compared with the N group, and hepatic PGC-1β mRNA expression was significantly increased in the HE group compared with the N and H group. Rapamycin administration alone did not affect PGC-1α, PGC-1β, PPARα, PPARβ, SREBP-1c, ChREBP, CPT1α, ACCα and PDK4 mRNA expression levels (Fig. 6) in fasting HFD rats with exercise or without exercise.
Figure 6.
Gene expression levels associated with energy metabolism in the liver tissues of rats fed N chow or HFD with or without E following R administration. Data are presented as the mean ± SE for n=4-6 rats. *P<0.05 vs. N group; #P<0.05 vs. H group. N, normal; H, HFD rats with sedentary; HFD, high-fed diet; E, exercise; R, rapamycin; PGC, peroxisome proliferator-activated receptor γ coactivator; PPAR, peroxisome proliferator-activated receptor; SREBP, sterol regulatory element binding protein; ChREBP, carbohydrate-response element-binding protein; CPT1α, carnitine palmitoyl transferase1α; ACCα, acyl-CoA carboxylase 1; PDK4, pyruvate dehydrogenase kinase 4.
Rapamycin inhibits hepatic mTORC1 activity
mTORC1 is sensitive, while mTORC2 is insensitive to the inhibition of their activity caused by rapamycin (5). However, it has been reported that long-term exposure to rapamycin can inhibit mTOR2 (25). To examine whether rapamycin could inhibit liver mTORC1 and mTORC2 activity, the phosphorylation levels of S6 (a marker of mTORC1 pathway activity) were measured. In addition, the phosphorylation of the ser473 site of Akt (a specific phosphorylation site of mTORC2) was assessed. Compared with the N group, the p-S6/S6 but not p-Akt/Akt ratio was significantly increased in the H group. Compared with the H group, the p-S6/S6 but not p-Akt/Akt ratio was significantly decreased in the HE, HR and HER groups. The results indicated that exercise and rapamycin significantly reduced the p-S6/S6 ratio without affecting p-Akt/Akt ratio in the liver tissues of HFD rats (Fig. 7).
Figure 7.
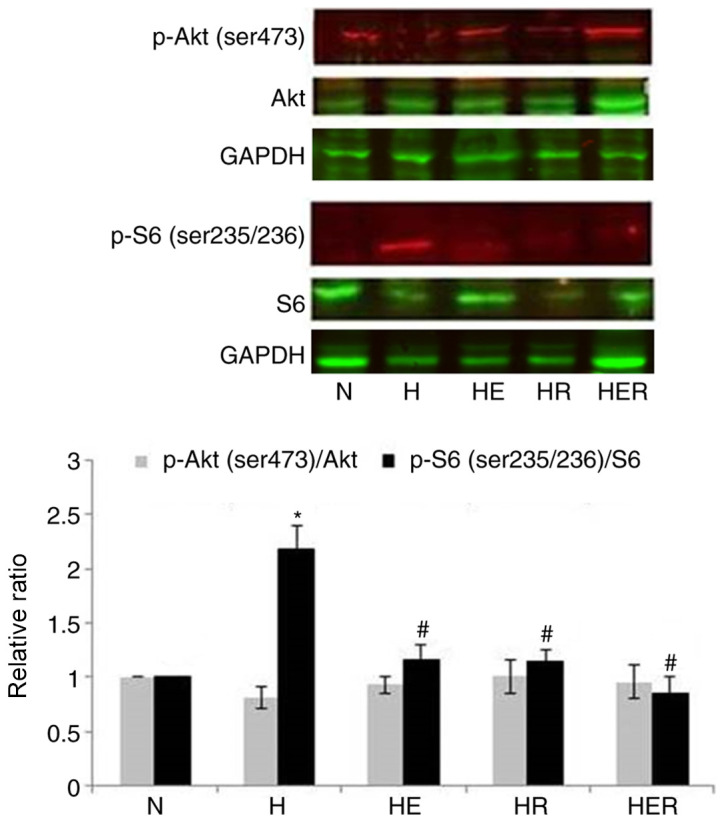
Effects of E and R administration on hepatic mTOR activity. Phosphorylation levels of S6 represent mTORC1 pathway activity, while the phosphorylation levels of the ser473 site of Akt represent mTORC2 pathway activity. Data are presented as the mean ± SE for n=4-6 rats. *P<0.05 vs. N group; #P<0.05 vs. H group. N, normal; H, HFD rats with sedentary; HFD, high-fed diet; E, exercise; R, rapamycin; p-, phosphorylated; S6, ribosomal protein S6; mTORC, rapamycin-sensitive mTOR complex.
Discussion
It has been reported that mTORC1 participates in the development of obesity-related insulin resistance. For instance, mTORC1 is overactivated in the liver of obese rats (26), while genetic inactivation of mTORC1 activity or acute rapamycin administration improves insulin sensitivity and alleviates hyperglycemia (19,27). However, chronic rapamycin administration (>2 weeks) may induce a diabetic-like syndrome, such as insulin resistance, glucose intolerance and dyslipidemia (16). In contrast to these findings, it has been shown that long-term administration of rapamycin exerts beneficial metabolic effects in diet-induced obese mice (28). Previous studies have also reported different effects of rapamycin on insulin sensitivity depending on the dose, route, frequency and time period of rapamycin administration (24,29–31). Moreover, the status of islet function is an important factor affecting insulin sensitivity caused by rapamycin in experimental animal models (32). Prolonged treatment with rapamycin results in the inactivation of both mTORC1 and mTORC2, which may be a cause of rapamycin-aggravated glucose intolerance and insulin resistance (25). The results of present study were in line with previous reports (18,19,33,34), demonstrating that mTORC1 was overactivated in the liver tissues of HFD rats and that both exercise and rapamycin treatment (2 weeks) effectively reduced hepatic mTORC1 activity without altering basal hepatic mTORC2 activity in HFD rats. It was found that rapamycin administration protected against obesity without affecting biochemical parameters including FBG, FFA, FTG and FINS, whereas it aggravated glucose intolerance and failed to suppress hepatic lipid deposition in HFD rats. Furthermore, exercise reduced visceral fat/body weight and hepatic lipid deposition, as well as improved glucose intolerance and insulin intolerance. Exercise combined with rapamycin administration also reduced body weight and visceral fat in HFD rats, while rapamycin impaired the exercise-induced improvement of glucose intolerance and insulin intolerance.
Mitochondrial dysfunction is an important cause of insulin resistance and of the development of metabolic disorders (35). Previous studies have reported that the decreased activity and increased acetylation of CPT1α, along with the repression of PPARγ activity by tafazzin, impair mitochondrial size and function, resulting in the development of insulin resistance and metabolic dysregulation (11,36). mTOR is also essential for maintaining mitochondrial function and energy balance (37,38). The present results demonstrated that rapamycin did not affect hepatic HADH activity (fatty acids β-oxidation), SDH activity and the mRNA expression levels of CPT1α, ACCα and PDK4, while it impaired exercise-induced hepatic COX activity (oxidative phosphorylation of mitochondria) in fasting HFD rats. The findings also indicated that rapamycin damaged mitochondrial oxidative capacity in HFD rats with or without exercise, which may contribute to rapamycin-aggravated glucose intolerance and insulin resistance, as well as counteract the effects of exercise on glucose tolerance and insulin tolerance.
In mammalian systems, PPARs, PGC-1α and PGC-1β serve critical roles in the modulation of mitochondrial oxidation (39–41), while transcription factors, such as SREBP-1c and ChREBP drive hepatic lipogenesis (42,43). PPARα is the key regulator of hepatic fatty acid β-oxidation and PPARβ indirectly modulates hepatic gluconeogenesis by regulating the expression of forkhead box-containing protein O subfamily-1 (39,42). PGC-1α induces hepatic gluconeogenesis, whereas PGC-1β is restricted to the maintenance of basal mitochondrial function and exerts dual roles in regulating fatty acid metabolism (40,44). The results of the present study indicated that HFD downregulated hepatic PPARβ gene expression, which may favor hepatic gluconeogenesis. In addition, exercise upregulated PGC-1α and PGC-1β mRNA expression levels in HFD rats, which may contribute to the reduction of liver TG levels via gluconeogenesis and fatty acid metabolism (44).
It has been reported that mTOR is critical in the regulation of lipid and glucose metabolism in the liver (12,14,44,45). For instance, inhibition of hepatic mTORC1 activity is required for fasting-induced hepatic PGC-1α and PPARα upregulation (14). In addition, mTORC1 upregulates SREBP-1c and ChREBP transcriptional activity (12,45,46). Despite this evidence, the present results demonstrated that rapamycin administration alone caused no effects on fasting hepatic PGC-1α, PGC-1β, PPARα, PPARβ, SREBP-1c and ChREBP mRNA expression levels in HFD rats, which may be associated with the dosage and time course of rapamycin treatment. However, there are several other important genes for hepatic metabolism and adipogenesis, such as PPARγ and fatty acid binding protein 4 (FABP4). A previous study revealed that the decreased activity of PPARγ led to insulin resistance and metabolic dysregulation (11). It has also been shown that high-intensity exercise significantly increases serum concentrations of FABP4 secreted by adipocytes via β-adrenergic mediated lipolysis (47). Moreover, adiponectin supplementation in pregnant mice prevents maternal obesity-associated fetal overgrowth by improving insulin resistance and restoring normal mTORC1 signaling (48). Therefore, the lack of detection of these genes in the model established in the present study was a limitation.
In conclusion, the present results indicated that chronic rapamycin administration reduced COX activity, while it exhibited no effects on the reduction of TG levels following exercise and on the upregulation of the mitochondrial metabolic gene expression in the liver tissues of HFD rats. The results suggested that exercise reduced TG content and upregulated mitochondrial metabolic gene expression in the liver of HFD rats, which may not be mediated via the mTOR pathway.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was supported by Natural Science Foundation of China (grant no. 31571217).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
BL conceived and designed the present study. BL, GT, CD, HQ and YW conducted the experiments and analyzed the data. BL and GT wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All the experimental procedures were approved by the Institutional Animal Care and Use Committee of the Guangzhou Sport University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lara-Castro C, Garvey WT. Intracellular lipid accumulation in liver and muscle and the insulin resistance syndrome. Endocrinol Metab Clin North Am. 2008;37:2932–856. doi: 10.1016/j.ecl.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuel VT, Shulman GI. Mechanisms for insulin resistance: Common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Meo S, Iossa S, Venditti P. Improvement of obesity-linked skeletal muscle insulin resistance by strength and endurance training. J Endocrinol. 2017;234:R159–R181. doi: 10.1530/JOE-17-0186. [DOI] [PubMed] [Google Scholar]
- 4.Golabi P, Locklear CT, Austin P, Afdhal S, Byrns M, Gerber L, Younossi ZM. Effectiveness of exercise in hepatic fat mobilization in non-alcoholic fatty liver disease: Systematic review. World J Gastroenterol. 2016;22:6318–6327. doi: 10.3748/wjg.v22.i27.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillén C, Benito M. mTORC1 overactivation as a key aging factor in the progression to type 2 diabetes mellitus. Front Endocrinol (Lausanne) 2018;9:621. doi: 10.3389/fendo.2018.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lushchak O, Strilbytska O, Piskovatska V, Storey KB, Koliada A, Vaiserman A. The role of the TOR pathway in mediating the link between nutrition and longevity. Mech Ageing Dev. 2017;164:127–138. doi: 10.1016/j.mad.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Kimball SR. Integration of signals generated by nutrients, hormones, and exercise in skeletal muscle. Am J Clin Nutr. 2014;99:237S–242S. doi: 10.3945/ajcn.113.068387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Kim SG, Blenis J. Rapamycin: One drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabatini DM. Twenty-five years of mTOR: Uncovering the link from nutrients to growth. Proc Natl Acad Sci USA. 2017;114:11818–11825. doi: 10.1073/pnas.1716173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Ouarrat D, Isaac R, Lee YS, Oh DY, Wollam J, Lackey D, Riopel M, Bandyopadhyay G, Seo JB, Sampath-Kumar R, Olefsky JM. TAZ Is a negative regulator of PPARgamma activity in adipocytes and TAZ deletion improves insulin sensitivity and glucose tolerance. Cell Metab. 2020;31:162–173 e165. doi: 10.1016/j.cmet.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci USA. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fletcher JA, Meers GM, Linden MA, Kearney ML, Morris EM, Thyfault JP, Rector RS. Impact of various exercise modalities on hepatic mitochondrial function. Med Sci Sports Exerc. 2014;46:1089–1097. doi: 10.1249/MSS.0000000000000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houde VP, Brûlé S, Festuccia WT, Blanchard PG, Bellmann K, Deshaies Y, Marette A. Chronic rapamycin treatment causes glucose intolerance and hyperlipidemia by upregulating hepatic gluconeogenesis and impairing lipid deposition in adipose tissue. Diabetes. 2010;59:1338–1348. doi: 10.2337/db09-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caron A, Mouchiroud M, Gautier N, Labbé SM, Villot R, Turcotte L, Secco B, Lamoureux G, Shum M, Gélinas Y, et al. Loss of hepatic DEPTOR alters the metabolic transition to fasting. Mol Metab. 2017;6:447–458. doi: 10.1016/j.molmet.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kucejova B, Duarte J, Satapati S, Fu X, Ilkayeva O, Newgard CB, Brugarolas J, Burgess SC. Hepatic mTORC1 opposes impaired insulin action to control mitochondrial metabolism in obesity. Cell Rep. 2016;16:508–519. doi: 10.1016/j.celrep.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenerson HL, Subramanian S, McIntyre R, Kazami M, Yeung RS. Livers with constitutive mTORC1 activity resist steatosis independent of feedback suppression of Akt. PLoS One. 2015;10:e0117000. doi: 10.1371/journal.pone.0117000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao B, Xu Y. Exercise improves skeletal muscle insulin resistance without reduced basal mTOR/S6K1 signaling in rats fed a high-fat diet. Eur J Appl Physiol. 2011;111:2743–2752. doi: 10.1007/s00421-011-1892-5. [DOI] [PubMed] [Google Scholar]
- 21.Bass A, Brdiczka D, Eyer P, Hofer S, Pette D. Metabolic differentiation of distinct muscle types at the level of enzymatic organization. Eur J Biochem. 1969;10:198–206. doi: 10.1111/j.1432-1033.1969.tb00674.x. [DOI] [PubMed] [Google Scholar]
- 22.Osumi T, Hashimoto T. Occurrence of two 3-hydroxyacyl-CoA dehydrogenases in rat liver. Biochim Biophys Acta. 1979;574:258–267. doi: 10.1016/0005-2760(79)90007-9. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Leontieva OV, Paszkiewicz GM, Blagosklonny MV. Comparison of rapamycin schedules in mice on high-fat diet. Cell Cycle. 2014;13:3350–3356. doi: 10.4161/15384101.2014.970491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khamzina L, Veilleux A, Bergeron S, Marette A. Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: Possible involvement in obesity-linked insulin resistance. Endocrinology. 2005;146:1473–1481. doi: 10.1210/en.2004-0921. [DOI] [PubMed] [Google Scholar]
- 27.Volkers M, Doroudgar S, Nguyen N, Konstandin MH, Quijada P, Din S, Ornelas L, Thuerauf DJ, Gude N, Friedrich K, et al. PRAS40 prevents development of diabetic cardiomyopathy and improves hepatic insulin sensitivity in obesity. EMBO Mol Med. 2014;6:57–65. doi: 10.1002/emmm.201303183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makki K, Taront S, Molendi-Coste O, Bouchaert E, Neve B, Eury E, Lobbens S, Labalette M, Duez H, Staels B, et al. Beneficial metabolic effects of rapamycin are associated with enhanced regulatory cells in diet-induced obese mice. PLoS One. 2014;9:e92684. doi: 10.1371/journal.pone.0092684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salmon AB. About-face on the metabolic side effects of rapamycin. Oncotarget. 2015;6:2585–2586. doi: 10.18632/oncotarget.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Diaz V, Fernandez E, Strong R, Ye L, Baur JA, Lamming DW, Richardson A, Salmon AB. Rapamycin-induced metabolic defects are reversible in both lean and obese mice. Aging (Albany NY) 2014;6:742–754. doi: 10.18632/aging.100688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang Y, Westbrook R, Hill C, Boparai RK, Arum O, Spong A, Wang F, Javors MA, Chen J, Sun LY, Bartke A. Duration of rapamycin treatment has differential effects on metabolism in mice. Cell Metab. 2013;17:456–462. doi: 10.1016/j.cmet.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reifsnyder PC, Flurkey K, Te A, Harrison DE. Rapamycin treatment benefits glucose metabolism in mouse models of type 2 diabetes. Aging (Albany NY) 2016;8:3120–3130. doi: 10.18632/aging.101117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenerson HL, Yeh MM, Yeung RS. Tuberous sclerosis complex-1 deficiency attenuates diet-induced hepatic lipid accumulation. PLoS One. 2011;6:e18075. doi: 10.1371/journal.pone.0018075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Binsch C, Jelenik T, Pfitzer A, Dille M, Müller-Lühlhoff S, Hartwig S, Karpinski S, Lehr S, Kabra DG, Chadt A, et al. Absence of the kinase S6k1 mimics the effect of chronic endurance exercise on glucose tolerance and muscle oxidative stress. Mol Metab. 2017;6:1443–1453. doi: 10.1016/j.molmet.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res. 2008;102:401–414. doi: 10.1161/CIRCRESAHA.107.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Softic S, Meyer JG, Wang GX, Gupta MK, Batista TM, Lauritzen HPMM, Fujisaka S, Serra D, Herrero L, Willoughby J, et al. Dietary sugars alter hepatic fatty acid oxidation via transcriptional and post-translational modifications of mitochondrial proteins. Cell Metab. 2019;30:735–753 e734. doi: 10.1016/j.cmet.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramanathan A, Schreiber SL. Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci USA. 2009;106:22229–22232. doi: 10.1073/pnas.0912074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 39.Han L, Shen WJ, Bittner S, Kraemer FB, Azhar S. PPARs: Regulators of metabolism and as therapeutic targets in cardiovascular disease. Part II: PPAR-b/δ and PPAR-g. Future Cardiol. 2017;13:279–296. doi: 10.2217/fca-2017-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villena JA. New insights into PGC-1 coactivators: Redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 2015;282:647–672. doi: 10.1111/febs.13175. [DOI] [PubMed] [Google Scholar]
- 41.Souza-Mello V. Peroxisome proliferator-activated receptors as targets to treat non-alcoholic fatty liver disease. World J Hepatol. 2015;7:1012–1019. doi: 10.4254/wjh.v7.i8.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimano H, Sato R. SREBP-regulated lipid metabolism: Convergent physiology-divergent pathophysiology. Nat Rev Endocrinol. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Viscarra J, Kim SJ, Sul HS. Transcriptional regulation of hepatic lipogenesis. Nat Rev Mol Cell Biol. 2015;16:678–689. doi: 10.1038/nrm4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chambers KT, Chen Z, Crawford PA, Fu X, Burgess SC, Lai L, Leone TC, Kelly DP, Finck BN. Liver-specific PGC-1beta deficiency leads to impaired mitochondrial function and lipogenic response to fasting-refeeding. PLoS One. 2012;7:e52645. doi: 10.1371/journal.pone.0052645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chau GC, Im DU, Kang TM, Bae JM, Kim W, Pyo S, Moon EY, Um SH. mTOR controls ChREBP transcriptional activity and pancreatic β cell survival under diabetic stress. J Cell Biol. 2017;216:2091–2105. doi: 10.1083/jcb.201701085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iizuka K. Recent progress on the role of ChREBP in glucose and lipid metabolism. Endocrine J. 2013;60:543–555. doi: 10.1507/endocrj.EJ13-0121. [DOI] [PubMed] [Google Scholar]
- 47.Iso T, Sunaga H, Matsui H, Kasama S, Oshima N, Haruyama Furukawa N, Nakajima K, Machida T, Murakami M, et al. Serum levels of fatty acid binding protein 4 and fat metabolic markers in relation to catecholamines following exercise. Clin Biochem. 2017;50:896–902. doi: 10.1016/j.clinbiochem.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 48.Aye IL, Rosario FJ, Powell TL, Jansson T. Adiponectin supplementation in pregnant mice prevents the adverse effects of maternal obesity on placental function and fetal growth. Proc Natl Acad Sci USA. 2015;112:12858–12863. doi: 10.1073/pnas.1515484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.