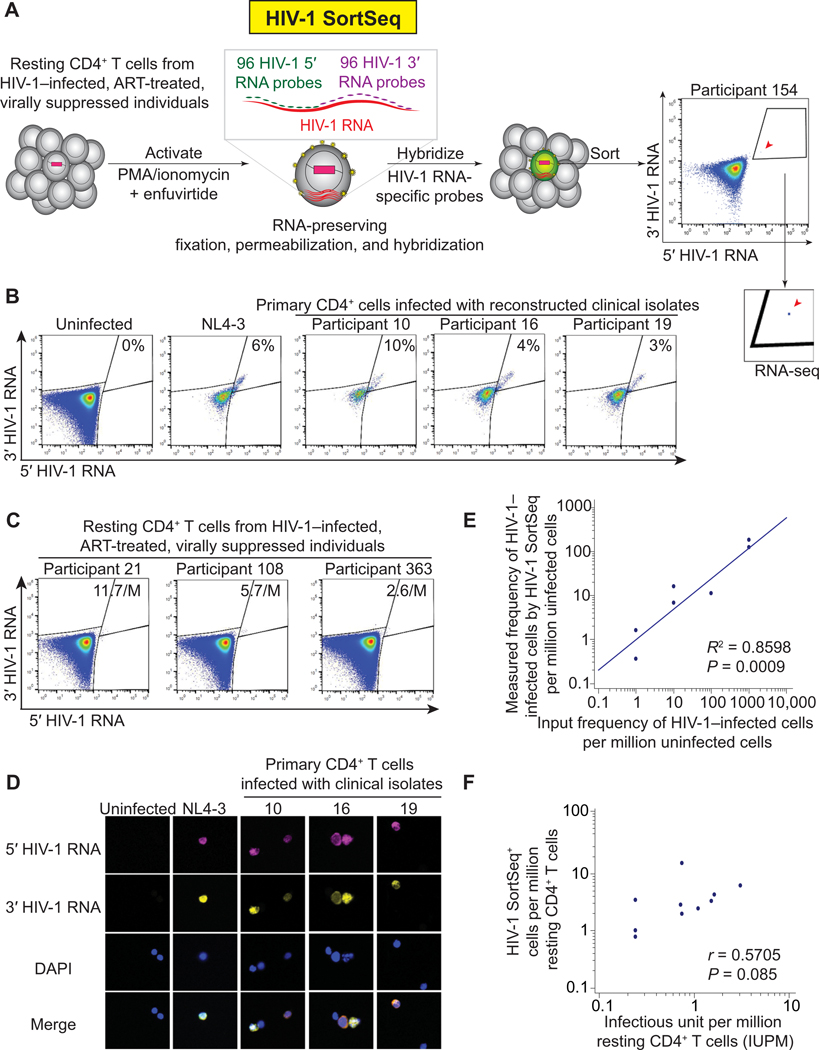
Fig. 1. HIV-1 SortSeq identifies CD4+ T cells harboring inducible HIV-1 from ART-treated, virally suppressed, HIV-1–infected individuals.
(A) Experimental scheme of HIV-1 SortSeq. CD4+ T cells from ART-treated, virally suppressed, HIV-1–infected individuals were sorted two-way into HIV-1 SortSeq+ and SortSeq− single cells. A stringent gating strategy away from the HIV-1 SortSeq− cells was used. (B) HIV-1 SortSeq gating strategy for primary cells infected with clinical isolates. The gating strategy was used to demonstrate the four quadrants of HIV-1 5’ and 3’ positive staining, not for sorting. (C) HIV-1 SortSeq gating strategy for CD4+ T cells isolated from HIV-1–infected, ART-treated, and virally suppressed individuals upon latency reversal. The gating strategy was used to demonstrate the four quadrants of HIV-1 5’ and 3’ staining, not for sorting. (D) Fluorescent microscopic imaging of primary CD4+ T cells infected with the NL4–3 reference strain and reconstructed clinical isolates at ~10% of infectivity. DAPI, 4,6-diamidino2-phenylindole nuclear staining. (E) Regression analysis of the frequency of HIV-1–infected cells detected by HIV-1 SortSeq versus the frequency of input HIV-1–infected cells. (F) Correlation between the frequency of HIV-1 SortSeq+ cells and the size of the latent reservoir as measured by viral outgrowth assays.