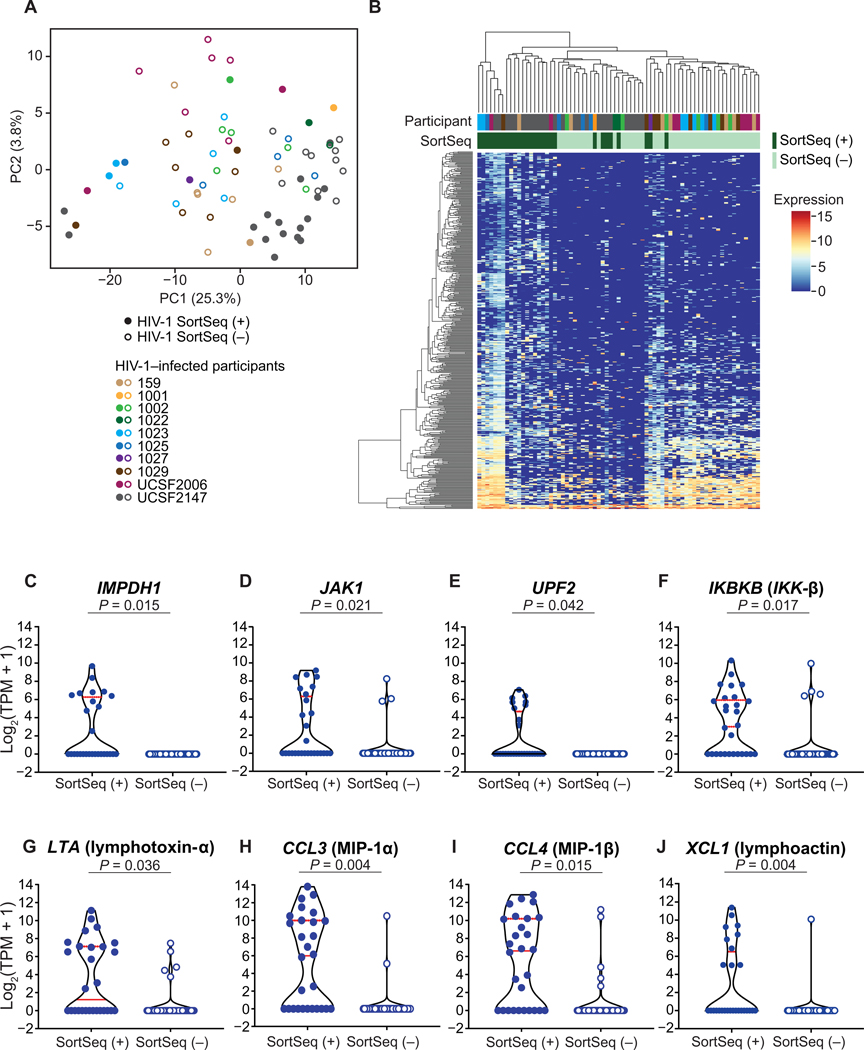
Fig. 3. The single-cell transcriptional landscape of HIV-1–infected cells upon ex vivo activation.
(A) The principal component analysis (PCA) demonstrates the distribution of HIV-1 SortSeq+ (closed circles) and SortSeq− cells (open circles) from ART-treated, virally suppressed, HIV-1–infected individuals. (B) Heterogeneous transcriptional profile of HIV-1–infected cells within 24 hours of latency reversal. The heat map shows significantly differentially expressed genes between HIV-1 SortSeq+ and SortSeq− cells. Values indicate expression measured as log2(TPM + 1). The participant ID is color-coded as shown in (A). ( C to J) Expression of selected significantly differentially expressed genes in HIV-1 SortSeq single cells. Each dot represents a single cell from 28 SortSeq+ and 43 SortSeq− cells. Red lines denote median expression. PC, principal component.