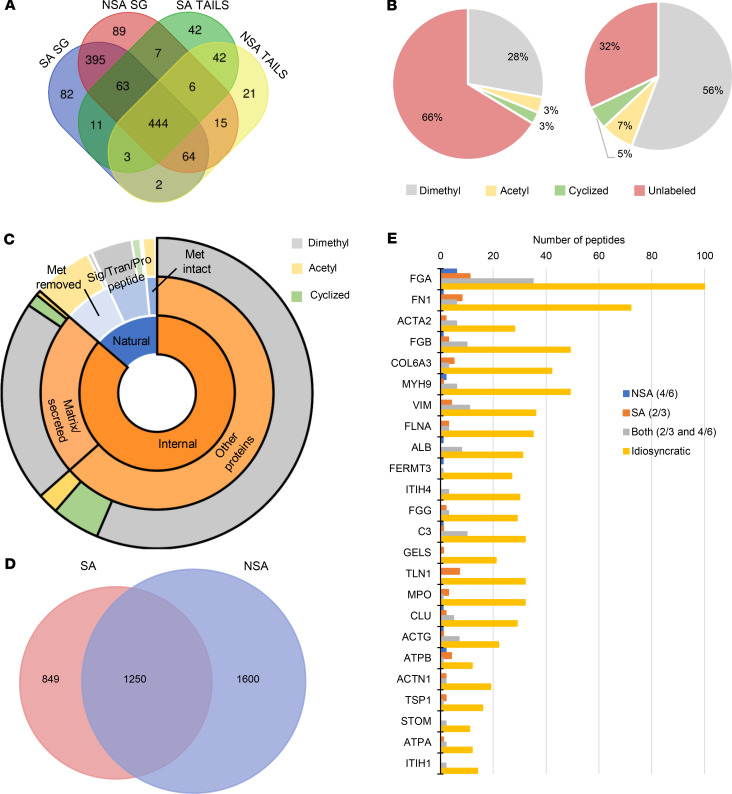
Figure 4. Overview of the vegetation proteome and degradome defined by TAILS.
(A) Venn diagram comparing proteins identified by at least 2 peptides in staphylococcal (SA) or non-staphylococcal (NSA) TAILS or single-run shotgun (SG) approaches. (B) Pie charts of peptides from the shotgun (left) and TAILS (right) with blocked N-termini (dimethyl, acetyl, or pyroglutamate modification as indicated in the key) or tryptic (unlabeled/unblocked) peptides. Blocked N-termini were enriched over 2-fold by TAILS. (C) Sunburst plot showing that blocked natural N-termini (blue shades) were preferentially acetylated (yellow, outer ring) or arose from constitutive N-terminal cleavage. These were not included in the degradome. Internal blocked peptides (orange) are typically blocked by dimethyl labeling (gray, outer ring) or cyclization (green, outer ring) but rarely acetylation (yellow, outer ring). (D) Venn diagram of TAILS-identified internal N-termini in SA and NSA groups. (E) Internal peptides seen in at least 4 of the NSA vegetations and none of the SA vegetations, in at least 2 of the SA vegetations and none of the NSA, or in at least 2 of the SA and 4 of the NSA (Both; 2 and 4), were compared with those seen inconsistently within groups (idiosyncratic).