Figure 2.
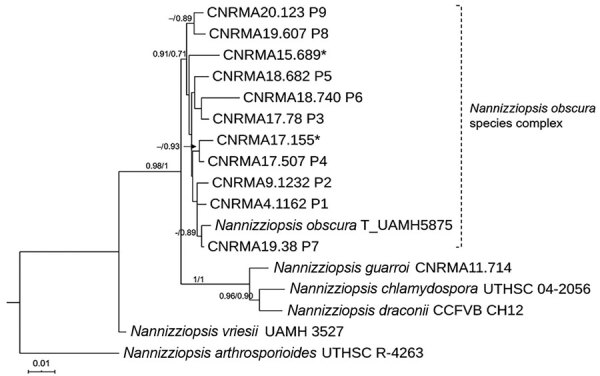
Maximum-likelihood tree obtained from combined large subunit ribosomal DNA, actin, and internal transcribed spacer 2 sequence data obtained from genomic analysis of Nannizziopsis obscura isolates from 9 patients from West Africa, France, 2004–2020, and reference sequences. Neighbor-joining bootstrap values or maximum-likelihood values are indicated on the branches. Support branch values <70% are not shown. Culture collection numbers appear next to sequences retrieved from GenBank, and type strains are indicated by a T after the species name. Patients from whom clinical isolates analyzed in this study were obtained are shown as P1–P9. The 2 isolates from patient 8 were morphologically and molecularly identical. Sequences marked with asterisks (*) refer to strains published by Nourrisson et al. (6). Scale bar indicates nucleotide substitutions per character.
