Figure 4: Methylation association with the androgen response pathway.
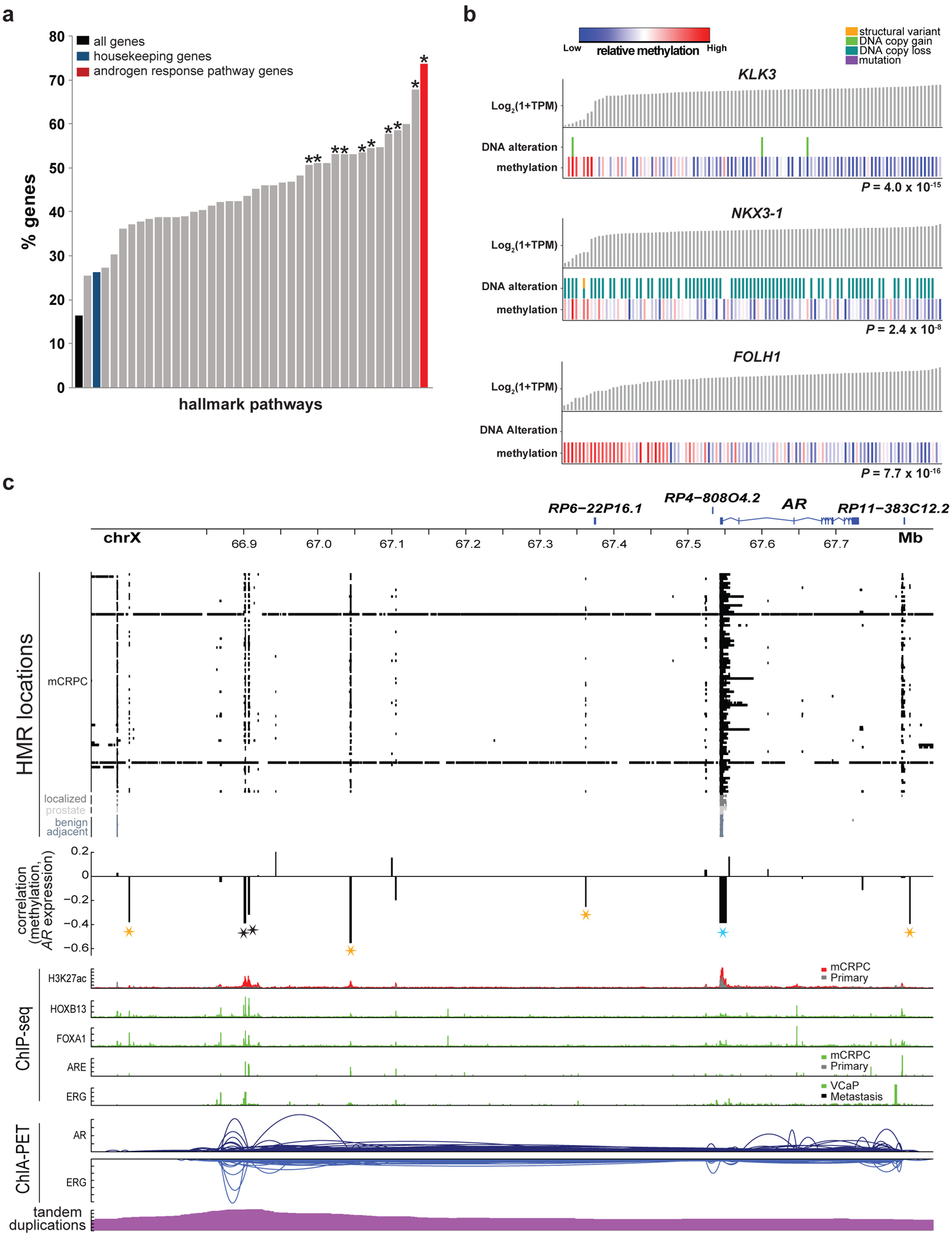
a, Percentage of genes in MSigDB Hallmark pathways for which methylation predicted expression independently from DNA alterations in a linear model. An asterisk indicates significant enrichment (two-sided FDR ≤ 0.05) relative to the set of all housekeeping genes. Significance was assessed with a two-sided Fisher’s exact test. N=100 independent mCRPC samples.
b, Sample-level gene expression levels compared to the presence of DNA alterations and methylation at the most significant promoter/gene body eHMR. Alterations predicted to be activating (KLK3, FOLH1) or inactivating (NKX3–1) are shown17). Significance of methylation levels was assessed by ANOVA comparing a model predicting gene expression from DNA alterations alone to a second model with methylation as an added factor, N=100 independent mCRPC samples.
c, HMRs, correlation between methylation in at loci harboring recurrent HMRs and AR expression, ChIP-seq peaks (H3K27ac33, AR36, ERG38, FOXA137, HOXB1337), and ChIA-PET interactions (AR and ERG)39 at the AR locus. Stars denote HMRs at which methylation was associated with AR expression (eHMRs), colored black for previously reported AR upstream enhancer, blue for the AR promoter, gold for new putative AR regulatory regions. Significance was assessed with a two-sided Spearman’s correlation test, N=100 independent mCRPC samples. “Primary” in the ChIP-seq tracks indicates localized primary prostate cancer.
