Figure 4.
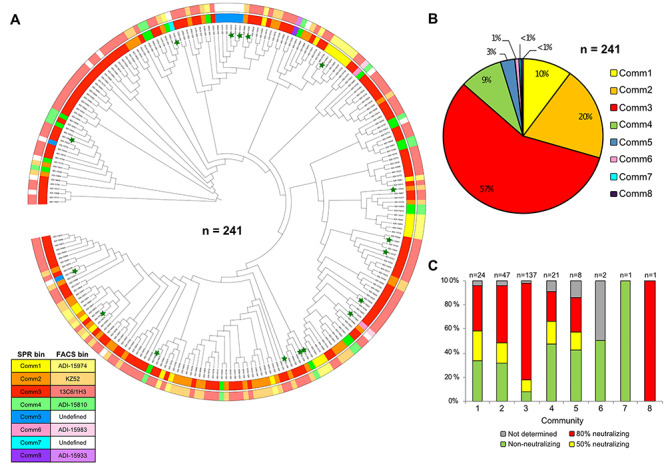
Benchmarking our HT-SPR epitope binning results against the literature. Antibody sequences, FACS binning and neutralization assay data were drawn from Bornholdt et al. [2]. (A) Sequence dendrogram of the antibody lineage of the variable heavy (VH) fragment alongside HT-SPR binning assignments (inner ring) and FACS binning assignments (outer ring) for the 241 antibodies that we empirically assigned to an epitope community. Antibody clones selected as pathfinders or unknowns to be tested in second pass binning are marked by a green star. Phylogenetic tree data generated by MUltiple Sequence Comparison by Log—Expectation (MUSCLE) [36], circular dendrogram figure constructed using Interactive Tree Of Life (iTOL) [37]. (B) Pie chart showing the distribution of 241 antibodies into the eight communities determined by HT-SPR. (C) Distribution of antibodies within each epitope community that reduced viral infectivity by 50, 80%, or determined to be nonneutralizing in a live virus plaque reduction neutralization assay. Neutralization assay was performed by and reported in Bornholdt et al. [2] as an endpoint titer based on the 50 and 80% thresholds in the number of plaques observed in control wells. The total number of antibodies within each assigned community is displayed at the top of each corresponding bar. Strongly neutralizing antibodies appear in all communities, except for Comm6 and Comm7, which were extremely rare.
