Figure 5:
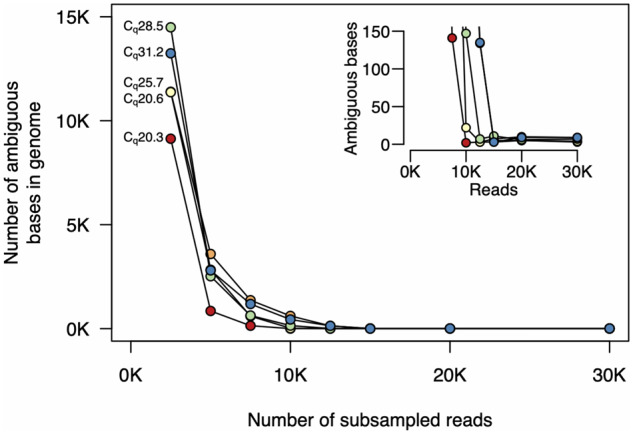
Numbers of ambiguous bases at different sequencing depths. We subsampled reads and used the filtering and assembly steps of the ARTIC Network bioinformatics pipeline. For all samples, <10 ambiguous bases remain after subsampling to 15 000 reads. For samples with lower Cq, only 10 000 reads are required. The inset plot shows higher resolution at the lower end of the y-axis. The colours of each sample on these plots are the same as those in Figs 3 and 4.
