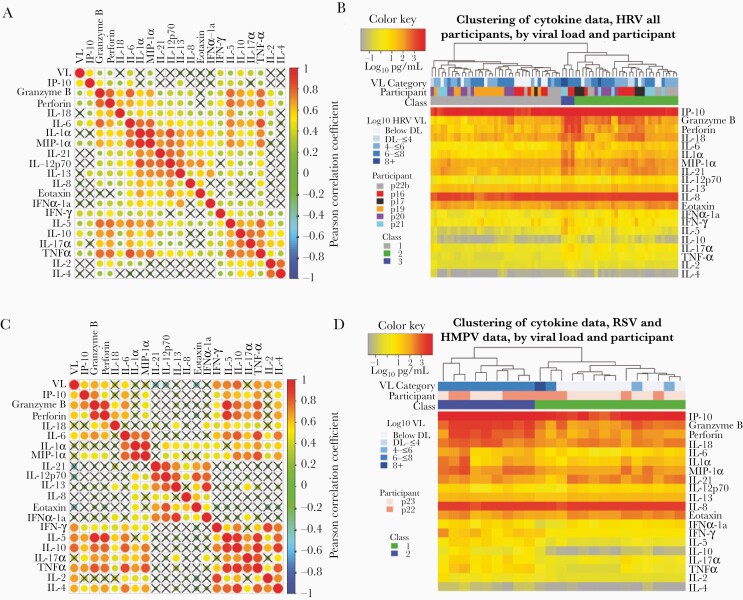
Figure 3.
Cytokines correlate according to cellular origin during respiratory virus infection, whereas samples cluster according to level of inflammation. A and B, Data from participants p16, p17, p18, p19, p20, p21, and p22b who have human rhinovirus (HRV) infection. C and D, Data from participants p22 and p23 who have respiratory syncytial virus (RSV) and human metapneumovirus (HMPV), respectively. A and C, Correlation plots with strong correlation according to cell type origin. X indicates a nonsignificant correlation. Color intensity and the size of the dot are proportional to the Pearson correlation coefficient. For both datasets, strong positive correlations are noted within cytokines linked with cytolytic T-cell responses, macrophage responses, and Th2 responses. B and D, Linkage clustering analysis of samples (columns) demonstrates classes of samples based on the concentration of inflammatory cytokines. B, For HRV infections, a minority of samples (class 3) from 2 participants and with the highest levels granzyme B, perforin, interleukin 6, interleukin 1α, macrophage inflammatory protein 1α, and interferon-γ all had high viral loads. All 6 participants had samples in the least inflammatory class (class 1) and 5 participants had samples in the moderate inflammatory class (class 2). D, For RSV and HMPV, inflammatory (class 2) and noninflammatory (class 1) sample clusters are evident. The inflammatory class of samples is highly associated with the highest viral loads. Abbreviations: DL, detection limit; HMPV, human metapneumovirus; HRV, human rhinovirus; IFN-α, interferon alpha; IFN-γ, interferon gamma; IL, interleukin; IP-10, interferon gamma-induced protein 10; MIP-1α, macrophage inflammatory protein 1 alpha; RSV, respiratory syncytial virus; TNF-α, tumor necrosis factor alpha; VL, viral load.