Abstract
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has shown once again that coronavirus (CoV) in animals are potential sources for epidemics in humans. Porcine deltacoronavirus (PDCoV) is an emerging enteropathogen of swine with a worldwide distribution. Here, we implemented and described an approach to analyze the epidemiology of PDCoV following its emergence in the pig population. We performed an integrated analysis of full genome sequence data from 21 newly sequenced viruses, along with comprehensive epidemiological surveillance data collected globally over the last 15 years. We found four distinct phylogenetic lineages of PDCoV, which differ in their geographic circulation patterns. Interestingly, we identified more frequent intra- and interlineage recombination and higher virus genetic diversity in the Chinese lineages compared with the USA lineage where pigs are raised in different farming systems and ecological environments. Most recombination breakpoints are located in the ORF1ab gene rather than in genes encoding structural proteins. We also identified five amino acids under positive selection in the spike protein suggesting a role for adaptive evolution. According to structural mapping, three positively selected sites are located in the N-terminal domain of the S1 subunit, which is the most likely involved in binding to a carbohydrate receptor, whereas the other two are located in or near the fusion peptide of the S2 subunit and thus might affect membrane fusion. Finally, our phylogeographic investigations highlighted notable South-North transmission as well as frequent long-distance dispersal events in China that could implicate human-mediated transmission. Our findings provide new insights into the evolution and dispersal of PDCoV that contribute to our understanding of the critical factors involved in CoVs emergence.
Keywords: porcine deltacoronavirus, evolution, recombination, phylogeographic, Bayesian inference, BEAST
Introduction
Coronaviruses (CoVs) are enveloped positive-stranded RNA viruses that are classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus belonging to the subfamily Orthocoronavirinae within the family of Coronaviridae and that exhibit a propensity for interspecies transmission. Betacoronaviruses can cause acute respiratory syndromes, such as severe acute respiratory syndrome (SARS)-CoV and Middle East respiratory syndrome coronavirus (MERS)-CoV (Drosten et al. 2003; Zaki et al. 2012), which are well-known for their ability to cross-species barriers and cause lethal respiratory infections in humans. In addition, HCoV-HKU1, HCoV-229E, HCoV-NL63, and HCoV-OC43 are not only endemic in humans but also of zoonotic origin (Corman et al. 2015, 2016; Su et al. 2016; Tao et al. 2017). Of note, the recent outbreak of Corona virus Disease 2019 (COVID-19), a cluster of pneumonia in humans caused by SARS-CoV-2, turned into a public health emergency of international concern (Sun et al. 2020). As of March 25, 2020, SARS-CoV-2 has infected 81,848 people in China and 414,179 globally (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports), causing thousands of deaths across the globe. Current research suggests that SARS-CoV-2 may have originated from bats directly or experienced adaptive evolution in an unknown intermediate host before transfer to humans (Zhou et al. 2020). The emergence and pandemic spread of SARS-CoV-2 represents a huge public health challenge and causes grave economic losses, emphasizing the importance of research dedicated to coronavirus cross-species transmission and spread in new host populations.
The high prevalence of CoV in mammals and birds may at least partly contribute to cross-species transmission dynamics of these viruses. The relatively high mutation and recombination rates ensure a large CoV genetic variability and can facilitate their adaptation to new hosts (Lau et al. 2018). A crucial step in cross-species transmission is the ability of such a virus to engage with new receptors in the novel host, which occurs through the spike (S) protein in CoVs (Li, Hulswit, et al. 2018).
An emerging Porcine deltacoronavirus (PDCoV) named HKU15 coronavirus was identified in pigs in Hong Kong in late 2012 (Woo et al. 2012). The virus infects the intestinal epithelia and causes acute, watery diarrhea, and vomiting with potentially fatal consequences (Ma et al. 2015). To date, PDCoV has been detected in at least 20 states within the United States (Li et al. 2014; Wang et al. 2014; Ma et al. 2015), as well as in Canada, South Korea (Lee S and Lee C 2014; Lee et al. 2016), China (Liu et al. 2017; Li, Feng, et al. 2018; Zhang, Liu, et al. 2019), Thailand (Madapong et al. 2016; Lorsirigool et al. 2017), Lao People’s Democratic Republic (Lorsirigool et al. 2016), Vietnam (Le et al. 2018), and Mexico (Perez-Rivera et al. 2019), posing a significant threat to the swine industry. So far, all other members of the Deltacoronavirus genus have been detected in birds (Saeng-Chuto et al. 2017), suggesting that birds are the natural host and ancestral reservoir of Deltacoronaviruses (Jung et al. 2015; Ma et al. 2015). Lau et al. demonstrated that the genome of PDCoV is closely related to the genome of sparrow CoV HKU17 and that it resulted from recombination between HKU17 and bulbul coronavirus HKU11 (Lau et al. 2018). Therefore, examining newly emerged CoVs causing epidemics in mammals provides opportunities for defining in detail the underlying events that allow viruses to cross the host range barrier and spread efficiently within new populations. Since swine are raised and traded for meat globally, their infection by CoVs and the close interaction with humans poses a threat to public health, as illustrated by the swine-origin human influenza virus outbreak (Smith et al. 2009). Although the prevention and control of coronavirus cross-species transmission and spread in the wildlife/livestock is critical, especially in China, detailed studies on the evolution and transmission of PDCoV are currently lacking despite the fact that recent years have seen the extensive use of phylogeographic approaches to unveil the dispersal history of viral epidemics.
To fill this gap, and as part of a nationwide swine virome metagenomics research project, we collected intestinal and fecal samples from swine farms in eastern and southern China where PDCoV outbreaks in 2018 and 2019 caused severe diarrhea and death in piglets. After performing virus isolation and next-generation sequencing analysis, we used full genome, S gene sequences and data from clinical diagnostic tests to examine the epidemiology and evolution of PDCoV during its emergence and spread among pigs, with an emphasis on the spread of the virus in China, the most endemic area of PDCoV and the country with the largest pig population worldwide. We aimed to unravel important aspects of PDCoV evolution and epidemiology, such as the source population, genetic recombination, time of origin, evolutionary rate, and the amino acid sites that might have played an important role in the adaptation of PDCoV to swine. Finally, we also employed phylogeographic approaches to examine the dispersal history and dynamics of emerging PDCoV in China.
Results
Sequencing, Virus Isolation, and PDCoV Epidemic in China
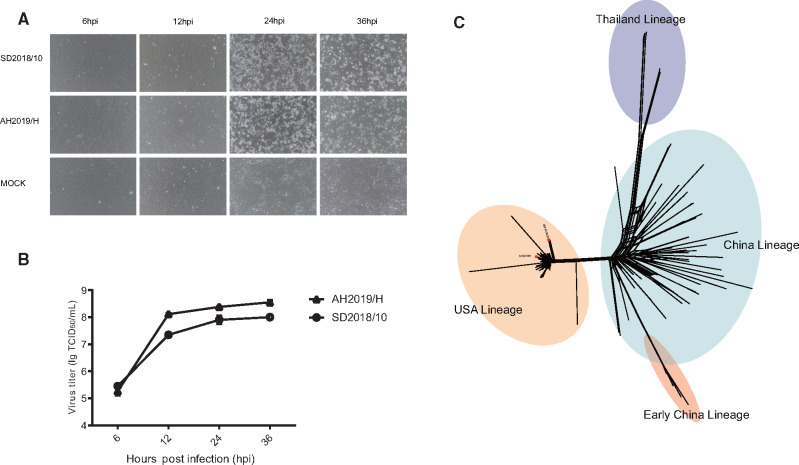
A total of 21 PDCoV full genomes and 29 S genes from seven provinces in China were sequenced. Two new PDCoV strains (named SD2018/10 and AH2019/H) were successfully isolated (fig. 1A and B and supplementary fig. S1, Supplementary Material online). Multistep replication curves revealed that the two PDCoVs had similar growth kinetics in LLC-PK1 cells, but the mean virus titer of AH2019/H was higher than the titer of SD2018/10 at 12, 24, and 36 hpi. Infected cells exhibited cytopathic effects, including shrinking, rounding, lighting, and disruptive morphological characteristics at 24 and 36 hpi (fig. 1A and B). Both viruses caused similar pathology in suckling pigs (He et al.; data not shown) .
Fig. 1.
Characteristics of isolated PDCoVs and phylogenetic analysis of full genomes. (A) Cytopathic effects and multistep replication curves of PDCoVs in LLC-PK1 cells at 6, 12, 24, and 36 hpi. Newly isolated viruses SD2018/10 and AH2019/H were used to infect LLC-PK1 cells at a MOI of 0.1. Magnification: 200×. (B) The viral titers (TCID50/ml) were determined at 6, 12, 24, and 36 hpi. The graph shows the mean of three different experiments. (C) Phylogeny reconstructed using 119 PDCoV genomes using the SplitsTree5 software with the Kimura 2-parameter model. Strains in blue regions correspond to the Thailand lineage, in red correspond to the Early China lineage, in orange to the USA lineage, and in green to regions of the China lineage.
The nucleotide similarity of the full genomes ranged from 98.9% to 99.8% among the newly sequenced samples and from 91.2% to 99.0% among reference sequences from NCBI. The nucleotide identity of the S gene of the newly sequenced samples ranged from 96.1% to 100%. The amino acid similarity among the newly sequenced samples ranged from 96.7% to 100% (supplementary table S3, Supplementary Material online). The phylogeny based on the full genomes revealed four major lineages, which we named the Thailand, Early China, USA, and China lineages (fig. 1C). The Thailand lineage actually contained strains from Vietnam, Laos, and Thailand. The Early China and China lineages contained strains only from China including the earliest strain that was isolated in the Anhui Province in 2004. The strains of the USA lineage circulated in the United States and spread to Japan, Korea, and China. Of note, two newly sequenced strains, AH2019/H and SD2019/426, clustered with the USA lineage (red dots in fig. 1C). This represents the first confirmed USA lineage strain in China.
PDCoVs Have Undergone Extensive Intra- and Interlineage Recombination in Their Evolutionary History
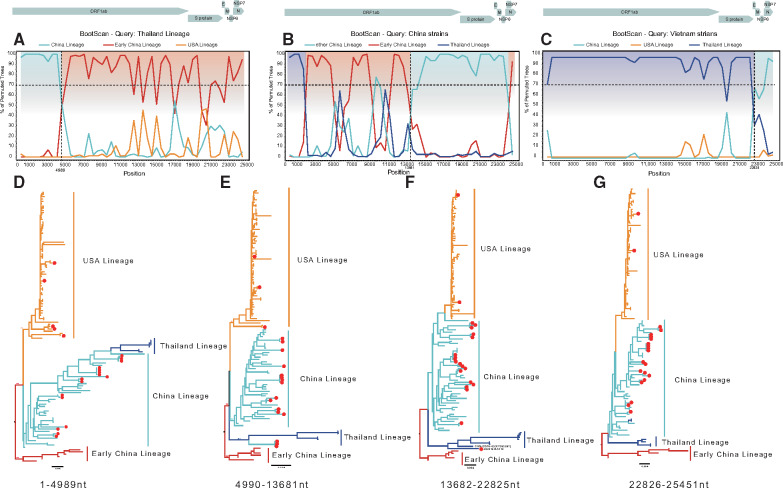
We used all full genome and S gene coding sequences for recombination analysis using RDP4. Half of these genome sequences were identified to be recombinant, with frequently shared breakpoints at nucleotide positions 4989 (fig. 2A), 13681 (fig. 2B), and 22825 (fig. 2C). We separated the genome sequences into four segments and performed Maximum likelihood (ML) phylogenetic reconstruction according to the breakpoints: 1–4989 (encoding ORF1a), 4990–13681 (encoding ORF1ab), 13682–22825 (encoding ORF1b and S), and 22826–25451 (encoding E + M + N) (fig. 2). Phylogenetic reconstruction and BootScan analysis indicated that the Thailand lineage viruses evolved from a recombinant virus that acquired the 5′ part (1–4989) of ORF1a from the China lineage (fig. 2A and D). In addition, some Chinese strains were closely related to American strains in this area of the genome (positions 1–4989). However, in the reconstructed phylogeny, the clustering of these subgenomic sequences was not supported by high bootstrap support values, suggesting that they have a more complex and uncertain history of recombination. We also found that two viruses from Vietnam in the Thailand lineage obtained the 3′ part containing the E, M, and N genes from the China lineage (fig. 2C and G). Moreover, some strains, for example, JS2019/A1414 (newly sequenced sample) and CHN-GD16-03 (KY363867), contained an S gene more similar to the Thailand lineage, with the other genes more closely related to the China lineage (fig. 2F). In addition, we found higher recombination rates in the China lineage as opposed to the USA lineage where no recombination was detected for full genome sequences (supplementary fig. S2, Supplementary Material online). We performed the same recombination analysis for the S gene. However, we only identified recombination events in 7.2% of the sequences.
Fig. 2.
Recombinant features of PDCoV phylogenies. (A–C) A rescaled structure of the PDCoV genome and bootscanning recombination analysis based on the variable genomic sites. The dashed line indicates 70% bootstrap support. The likely recombination region is shaded in a different color and colored broken lines represent different lineages: blue indicates the Thailand lineage, red the Early China lineage, orange the USA lineage, and green the China lineage. (A) Breakpoint at nucleotide 4989. (B) A breakpoint at nucleotide 13681. (C) A breakpoint at nucleotide 22826. (D–G) ML phylogenetic trees inferred for the different recombinant regions: nucleotides 1–4989, 4990–13681, 13682–22825, and 22826–26451. The ML tree was reconstructed using RAxML (v8.4.10) using a general time-reversible model accommodating among-site rate heterogeneity. In total, 1,000 bootstraps were evaluated to assess support values. The Thailand lineage is indicated in blue; the Early China lineage in red; the USA lineage in orange; and the China lineage in green. The red dots indicate strains that were sequenced as part of this study.
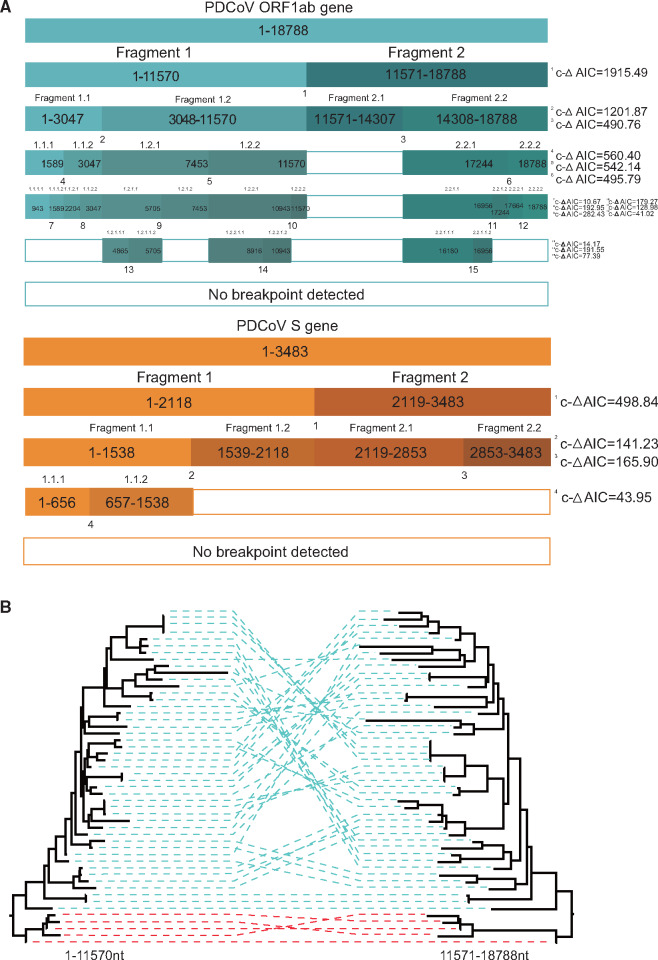
Using RDP4 analysis, we identified intralineage recombination hot spots of the China and Early China lineages sequences mainly located in the ORF1ab and S genes (data not shown). In order to further clarify the characteristics of recombination, we removed the interlineage recombination sequences in the China and Early China lineages. The GARD method was used to identify the intralineage breakpoints. The first breakpoint was the nucleotide position 11570 (corrected ΔAIC = 1,915.49), roughly at the end of the coding sequence of the ORF1a protein (fig. 3A). The two phylogenies inferred from sequences before and after this breakpoint were significantly incongruent (fig. 3B). A second analysis of the resulting fragment 1 (positions 1–11570) and fragment 2 (positions 11571–18788) yielded a further breakpoint in fragment 1 at position 3047 (corrected ΔAIC = 1,201.87), located in NSP2 and a further breakpoint in fragment 2 at position 14307 (corrected ΔAIC = 490.76) at the beginning of ORF1b. We repeated this procedure until no more breakpoints were identified (fig. 3A). There were 15 breakpoints in total in the ORF1ab gene. In the S gene, only four breakpoints were identified at nucleotide positions 656 (corrected ΔAIC = 43.95), 1538 (corrected ΔAIC = 141.23), 2118 (corrected ΔAIC = 498.84), and 2853 (corrected ΔAIC = 165.90). In summary, the recombination analyses of PDCoV revealed frequent recombination in the ORF1ab region (fig. 3A).
Fig. 3.
Frequent intralineage recombination events in China and Early China lineages. (A) Summary of GARD results. Colored boxes indicate fragments resulting from GARD-inferred breakpoints with corrected ΔAIC values shown on the right. (B) ML phylogenetic trees inferred for China and Early China lineage PDCoV ORF1ab using RAxML under the general time-reversible substitution model with gamma-distributed rates across sites. In total, 1,000 bootstraps were evaluated to assess support values. Trees were reconstructed for both regions separated by the breakpoint at position 11570.
Evolutionary Dynamics of PDCoV
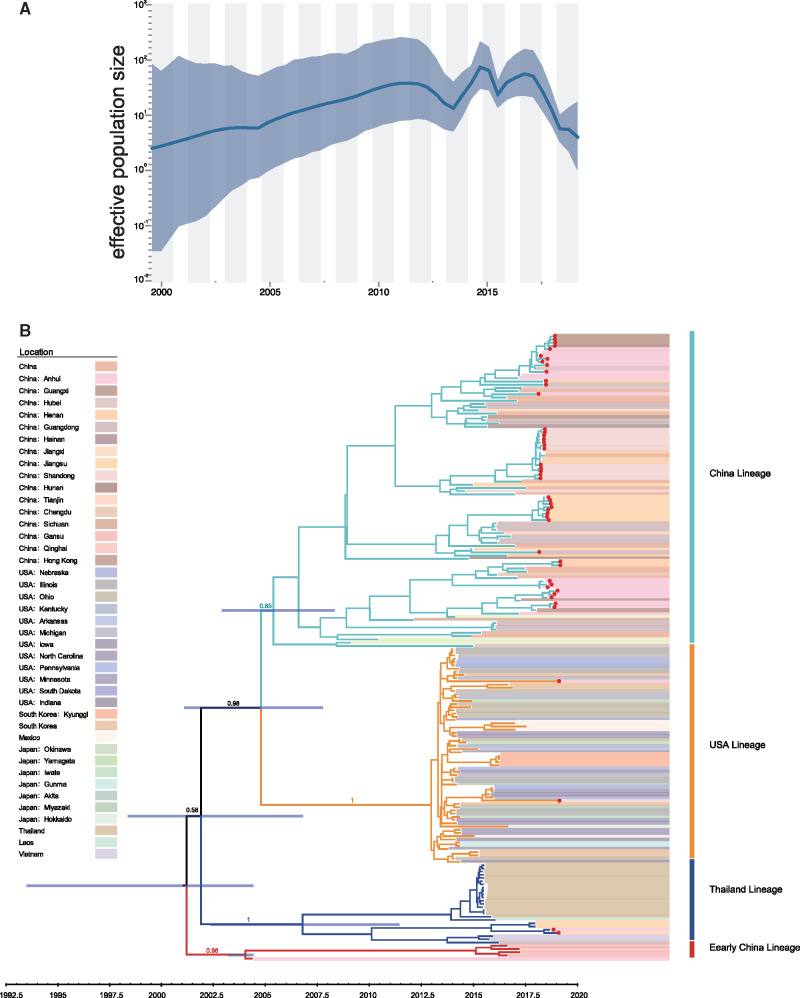
Since the number of sequences available for the S gene is larger than for full genome sequences, with relatively few mosaic genomes identified, we focused on these sequences for phylodynamic reconstruction. We first assessed the temporal signal in TempEst which revealed sufficiently strong temporal signal (R2 = 0.39) to estimate time-calibrated phylogenies using molecular clock models (supplementary fig. S3B, Supplementary Material online). Similar to the full genome, four lineages were observed according to the ML and maximum clade credibility (MCC) trees (supplementary figs. S3A and 4, Supplementary Material online). The MCC tree revealed the detection of all lineages of PDCoV in China (red dots in fig. 4B). In addition, our reconstruction confirmed that the virus spread from the United States to Mexico, as described in a previous study (Perez-Rivera et al. 2019), and observed that the USA lineage further spread to other countries, like China, Japan, and South Korea, through the hog trade (fig. 4). We estimated the time of the most recent common ancestor (tMRCA) of PDCoV at 1,999.5 with a 95% highest posterior density (HPD) range of (1,993.3–2,004.4). The estimated tMRCA was 2,006.8 (2,002.4–2,011.4) for the Thailand lineage, 2,004.4 (2,003.3–2,004.4) for the Early China lineage, 2,013.0 (2,012.6–2,013.3) for the USA lineage, and 2,005.4 (2,002.9–2,008.3) for the China lineage. The mean evolutionary rate was estimated at 1.70 (1.37–2.02) × 10−3 substitutions/site/year. As shown in supplementary figure S3C, Supplementary Material online, lineage-specific evolutionary rate estimates were 2.71 (0.08–6.92) × 10−3 subst/site/year for the Thailand lineage, 2.45 (0.02–5.20) × 10−3 subst/site/year for the Early China lineage, 1.21 (0.04–2.50) × 10−3 subst/site/year for the USA lineage, and 1.58 (0.10–4.21) × 10−3 subst/site/year for the China lineage. The estimated effective population sizes over time (fig. 4) show that PDCoV went through rapid expansion from its first occurrence in 2000 to 2011 after which the population size fluctuated at a relatively high level (fig. 4A).
Fig. 4.
Demographic history of PDCoV in pigs. (A) Demographic history inferred via a skygrid coalescent tree prior. The intervals represent 95% HPD of the product of generation time and effective population size Ne(t). The middle line tracks the inferred median of Ne(t). (B) MCC tree of the S gene constructed using BEAST (version 1.10.5) under the skygrid nonparametric coalescent model. The red dots indicate strains were sequenced in this study.
Protein Structure Analysis of Adaptive Evolution Sites
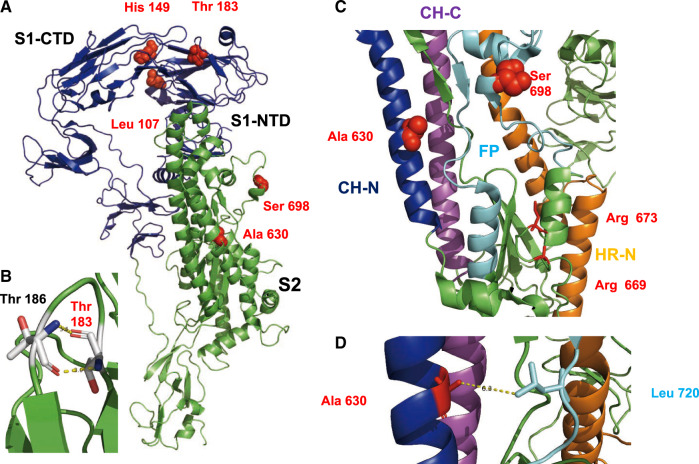
We identified five codons (sites 107, 149, 183, 630, and 698) under positive selection in the S protein (table 1). The identified sites were visualized in the Cryo-EM structure of S, which is a trimer composed of two subunits. The N-terminal S1 subunit (shown in blue in fig. 5A) contains the receptor binding domain, whereas the S2 subunit (shown in green) contains the membrane fusion activity. The S gene is the main determinant of host tropism and induces an antibody response. It is heavily N-glycosylated, but none of the exchanges would delete or create a glycosylation site, although it has been proposed that PDCoVs could evade immunity by glycan shielding (Xiong et al. 2018). Three amino acids under selection are located in S1 and two in S2. The amino acids in the S1 subunit, sites 183, 149, and 107, are located to the N-terminal domain (NTD). NTD adopts a β-sandwich fold that is identical to that of human galectin, a sugar-binding protein. Amino acid site 149, which shows the highest variability between lineages (supplementary table S4, Supplementary Material online), is present at the top of S (supplementary fig. S4, Supplementary Material online). Thr 183 is located in a loop between two β -sheets. Its main chain atoms form hydrogen bonds with Thr 186 which might stabilize the loop (fig. 5B).
Table 1.
Selection Analysis of the PDCoV S Protein.
| Site | FUBAR (Post.Pro) | MEME (P value) | FEL (P value) | SLAC (P value) |
|---|---|---|---|---|
| 107 | 0.979 | 0.02 | 0.015 | 0.026 |
| 149 | 0.998 | 0.01 | 0.004 | 0.027 |
| 183 | 0.968 | 0.04 | 0.027 | 0.089 |
| 630 | 0.98 | 0.00 | 0.012 | 0.007 |
| 698 | 0.99 | 0.02 | 0.012 | 0.087 |
Fig. 5.
Location of selected amino acids in the structure of S protein. (A) Cartoon representation of an S monomer. The S1 subunit is represented in blue and the S2 subunit in green. Selected amino acids are shown as red spheres. Leu 107 and His 149 are located in the N-terminal domain of S1 (S1-NTD) that binds to unidentified sugars, but close to the C-terminal domain of S1 (S1-CTD) that contains the binding site for the protein receptor. (B) Hydrogen bonds formed between Thr 183 and Thr 186 that might stabilize a loop. (C) Part of the structure of S2 with individual elements drawn in different colors. Central helix N (CH-N) is represented in blue, the central helix C (CH-C) in magenta, the fusion peptide (FP) in cyan, and the heptad repeat N in orange. Arg 669 and Arg 673, shown as red sticks, are presumed proteolytic cleavage sites. The selected amino acids Ala 630 and Ser 698 are shown as red spheres. (D) Detail of S2 showing the selected amino acid Ala 630 in proximity (6.3 Å) to Leu 720 in the fusion peptide.
The S2 subunit is composed of the central helix N (CH-N, blue in fig. 5C), a fusion peptide (FP, cyan), a heptad repeat region N (HR-N, orange), the central helix C (CH-C, magenta), and an unstructured heptad region C. S is activated by a trypsin-like protease at arginine 669 and 673. Acidic pH exposes the fusion peptide and HR-N and HR-C form the typical six helix bundle. One amino acid under selection, Ser 698, is located in a helix within the fusion peptide and exposed at the surface of the molecule. The other, Ala 630, is located deeper down in the interior of the trimeric spike in the central helix N (fig. 5C and supplementary fig. S4, Supplementary Material online).
Spatiotemporal Reconstruction of PDCoV Spread in China Using Discrete and Continuous Phylogeographic Approaches
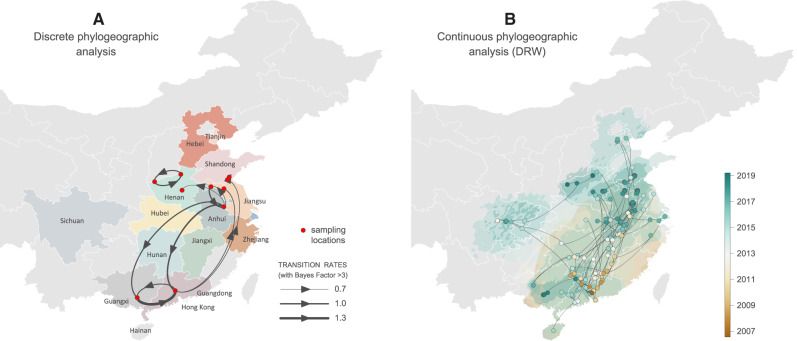
Using a generalized linear model (GLM) parameterization of the Bayesian discrete phylogeographic inference approach, we tested whether geographic distance and the average number of estimated annual live hogs in infected farms were predictive of the PDCoV patterns of spatial spread between the farms sampled in this study. This analysis did not provide any support for these predictors (Bayes Factor (BF) support <3), which could at least be partly explained by a lack of power due to the limited data size used for the analysis. When performing a regression of pairwise geographic distances against patristic distances computed on the MCC tree of the discrete-GLM analysis, we detected a significant but indeed weak correlation among the two distances (R2 ∼ 5%, P value < 0.001; supplementary fig. S5, Supplementary Material online). Different from the discrete-GLM inference, geographic distances within clusters of the same farm will contribute to this support for some degree of “isolation-by-distance.” To further uncover the PDCoV spatial diffusion patterns, we performed three distinct continuous phylogeographic analyses: one analysis with the relaxed random walk (RRW) and two analyses with the directional random walk (DRW) model, that is, a DRW analysis allowing both longitudinal and latitudinal drift as well as a DRW analysis only allowing a latitudinal drift. We estimated a significant latitudinal drift for both DRW analyses, that is, a mean latitudinal drift shift value of 0.48 with a 95% HPD interval (0.02–1.00) excluding 0 (hence rejecting a value of 0 that corresponds to an absence of drift). We therefore focused on the DRW analysis with latitudinal drift to summarize continuous phylogeographic patterns, even if associated with a higher phylogeographic uncertainty than the continuous phylogeographic reconstruction obtained under the RRW model (supplementary fig. S6, Supplementary Material online). Interestingly, the continuous phylogeographic reconstruction performed under the DRW model showed a southern origin of the lineage dispersal (fig. 6 and supplementary fig. S6, Supplementary Material online), which is consistent with epidemiological records pointing toward a China lineage of PDCoV origin nearby or in the Guangdong province (Woo et al. 2012). Furthermore, this continuous phylogeographic reconstruction highlights frequent long-distance dispersal events. For the selected DRW analysis, we estimated a notable weighted lineage dispersal velocity equal to 185 km/year (95% HPD [135–234]), whereas the RRW analysis estimates a considerably lower dispersal velocity (134 km/year, 95% HPD [98–179]).
Fig. 6.
Spatiotemporal diffusion of China PDCoV lineage within China as estimated from discrete (A) and continuous (B) phylogeographic reconstructions. (A) The discrete phylogeographic analysis was performed with the Bayesian stochastic search variable selection (BSSVS) approach, for which we displayed the intensity of the transition rates associated with a BF support higher than 3. (B) Continuous phylogeographic analyses were performed with the DRW diffusion model (only including a latitudinal drift; see the text for further details). For the continuous reconstruction, we mapped the MCC tree and 95% HPD regions based on trees subsampled from the post burn-in posterior distribution of trees. Nodes of the tree are colored according to a color scale ranging from brown (tMRCA) to green (most recent sampling time). Ninety-five percent highest posterior density (HPD) regions were computed for successive time layers, superimposed using the same color scale reflecting time cropped using Chinese international borders (see supplementary fig. S6, Supplementary Material online, for noncropped 95% HPD polygons as well as a comparison with the continuous phylogeographic reconstruction obtained under the relaxed random walk model). On both maps, subnational Chinese province borders are represented by white lines.
Discussion
PDCoV spread constitutes a risk for both the pig industry and public health because CoVs have repeatedly crossed the host barrier between different animals, like swine acute diarrhea syndrome coronavirus from bats to swine (Li, He, et al. 2018; Zhou et al. 2018; He et al. 2019) and from an animal reservoir to humans for the ongoing SARS-CoV-2 pandemic (Zhou et al. 2020). Here, we show that phylogenetically distinct lineages cocirculated globally, and that multiple inter- and intralineage recombination events are frequent in PDCoV, except in the USA lineage. We also report that the USA lineage virus was introduced to China around the winter of 2018. Both the USA and China lineages are the major prevailing genotypes worldwide. New lineages created by recombination events circulate in Thailand, Vietnam, and China.
Recombination increases virus genetic diversity and may contribute to adaptation to a new host (Woo et al. 2006; Lau et al. 2011; Huang et al. 2013; Sabir et al. 2016; Su et al. 2016; Forni et al. 2017). CoVs have a very high frequency of homologous RNA recombination as a result of random template switching during RNA replication thought to be mediated by a “copy choice” mechanism of the viral polymerase (Tao et al. 2017). In this study, we found high levels of recombination of PDCoV in the China, Early China, and Thailand lineages with multiple distinct genotypes. Moreover, in the China and Thailand lineages, recombination occurs frequently within ORF1ab which may therefore be a recombination hot spot in PDCoV similar to the S gene. Furthermore, most of the recombination events involved breakpoints in the ORF1a gene. This is specifically the case for the breakpoint at position 4989 located between nsp3 and nsp4 which encodes the polymerase. ORF1ab encodes the nonstructural proteins, including proteins involved in mitigating the innate immune response, for example nsp5, a protease that cleaves not only the viral polyprotein1a but also STAT 1, a signaling molecule involved in activating the type 1 interferon pathway (Zhu et al. 2017). Thus, one could speculate that recombination within ORF1a may contribute to the evasion of the swine innate immune system. In contrast, strains from the USA lineage do not exhibit recombination within the full-length genome and are associated with a relatively low diversity. In the United States, a higher level of biosecurity management reduces the risk of introducing viruses into farms or results in shorter virus outbreaks thereby reducing the risk for recombination events by reducing the probability of coinfections. Multiple pig production systems coexist in Asia and in China (Gilbert et al. 2015) allowing pigs to be traded through diverse value-chains which might explain higher rates of recombination. A previous study showed that recombination within or around the S gene is likely a common phenomenon among Deltacoronavirus and other Coronaviruses (Forni et al. 2017; Tao et al. 2017; Lau et al. 2018; Guo et al. 2019), which may facilitate interspecies transmission and adaptation to new animal hosts. Our results demonstrate that ORF1a recombination may also play an important role in the natural evolution of coronavirus among swine. In addition, the PDCoV S gene evolved at a mean rate of 1.67 × 10−3 subst/site/year over 15 years of evolution in pigs, which is significantly lower than described for another important swine coronavirus, porcine epidemic diarrhea virus (Sung et al. 2015). Long-term and widespread use of vaccines is likely to affect porcine epidemic diarrhea virus evolution and thus offers an explanation for the lower rate of nucleotide substitution in PDCoV because it is subject to weaker immune selection. Of note, our results also suggest that vaccine usage and different farming models and ecologic environment might have a great impact on recombination, evolution, and population diversity of emerging CoVs.
Our study provides the first phylogeographic exploration of PDCoV in China. Using continuous phylogeographic reconstruction, we estimated a relatively high weighted lineage dispersal velocity ranging from ∼134 km/year (RRW model) to ∼185 km/year (DRW model). These values are much higher than, for instance, estimates previously reported for rabies virus spread (Dellicour et al. 2017, 2019; Tian et al. 2018) but still lower than, for instance, the weighted branch dispersal velocity estimated for the West Nile virus spread in North America (255 km/year, 95% HPD [231, 286]) (Pybus et al. 2012). Although our discrete phylogeographic inference results currently do not implicate geographic distance or hog population sizes in farms as drivers of spatial spread, more comprehensive sampling and incorporating additional predictors at different levels (county, prefecture, or province) may elucidate important spatial dynamics in the future. It is possible that pig farm workers and contaminated feed could play a role in PDCoV dissemination, which may be difficult to formalize as predictors. Additionally, contamination is also likely to occur within a province or between neighboring provinces, whereas pig trade between provinces is deemed more likely. Wild animals in the vicinity of farms could also be implicated, but one would expect that this would to some extent be reflected in support for geographic distance. The continuous phylogeographic reconstruction that incorporates a directional tendency suggests a southern origin for PDCoV spread in China. Both discrete and continuous phylogeographic investigations highlight notable South-North transmission as well as frequent long-distance dispersal events in China that could implicate human-mediated transmission and which would be in line with the relatively high branch dispersal velocity estimated for this PDCoV spread.
Five codons (107, 149, 183, 630, and 698) were determined to be under positive selection. Ser 698 in S2 is located within the fusion peptide but is replaced by amino acids with similar properties (Ala, Thr). Ala 630 is located near the fusion peptide and if replaced by larger amino acids, such as Leu in the China lineage, might interact by hydrophobic forces with Leu 720 in the fusion peptide (fig. 5C and D). Thus, both amino acids might be involved in membrane fusion, for example, by affecting exposure of the fusion peptide. The other three residues under positive selection are located in the NTD of S1, which is a lectin (Shang et al. 2018). Interestingly, residue 149 shows the highest variability within lineages and it is replaced by amino acids with very different properties (supplementary table S4, Supplementary Material online). This site might modulate attachment to sugars which are used as attachment factors before S binds to aminopeptidase N via the C-terminal domain of S1 (S1-CTD) (Li, Hulswit, et al. 2018; Shang et al. 2018; Wang et al. 2018; Xiong et al. 2018) or be part of an epitope positively selected by pig antibodies.
In summary, assuming that PDCoV has relatively recently been introduced from birds into pigs, the amino acids identified under selection might contribute to better entry of PDCoV into pig and human cells (Li, Hulswit, et al. 2018), both by evolution toward more efficient receptor binding and by enhanced membrane fusion, that is, and thus could confer a fitness advantage to the virus. These results support the idea that evolution and transmission of CoVs upon emergence in new hosts involves a complex and variable series of steps. Continued surveillance studies in swine are needed to monitor the geographical spread and seasonal patterns of PDCoV and other CoVs infections as well as the detection of recombinant viruses.
Materials and Methods
Sample Collection and Virus Isolation
PDCoV-positive gut and feces samples were collected from ten intensive pig farms and one free-range household, located in the Anhui, Jiangsu, Henan, Zhejiang, Fujian, Guangxi, and Shandong provinces in China from 2018 to 2019. PDCoV isolation was performed using LLC-PK1 cells (ATCC CL-101) as reported previously (Hu et al. 2015). LLC-PK1 cells were grown in Dulbecco’s modified eagle medium (DMEM, Gibco, USA) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco), 1% (v/v) penicillin–streptomycin, and 4 mM l-glutamine. PDCoV isolation was confirmed by reverse transcription-polymerase chain reaction (supplementary table S1, Supplementary Material online) and an indirect immunofluorescence assay.
Genome and Spike Gene Sequencing
Virus RNAs were extracted using the TIANGEN viral RNA kit (TIANGEN, China). The cDNA was synthesized using random primers and the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA). Each sample was first screened by polymerase chain reaction followed by viral isolation or purification, sequenced by sanger or metagenomic sequencing using Hiseq2500 Illumina (Chong et al. 2019; Zhang, Cheng, et al. 2019).
Sequence Analysis
All available PDCoV genome sequences and S protein coding sequences up to June 2, 2019 were retrieved from NCBI GenBank (https://www.ncbi.nlm.nih.gov/). A total of 119 genome sequences and 220 S gene sequences were used, along with 21 newly sequenced genomes (GenBank accession numbers in supplementary table S2, Supplementary Material online) and 29 newly sequenced S gene (GenBank accession numbers: MN058045–MN058073) sequences, respectively. All sequences were aligned using MUSCLE and manually adjusted in MEGA7 (Edgar 2004; Kumar et al. 2016). Genome sequences were aligned at the nucleotide level, whereas the S protein coding sequences were aligned at the amino acid level with manual adjustments.
Recombination Analysis
Full genomes and S gene sequences were analyzed separately to detect recombination events using RDP4 (Martin et al. 2015). A total of seven methods implemented in RDP4 were applied including RDP (Martin and Rybicki 2000), GENECONV (Padidam et al. 1999), 3Seq (Boni et al. 2007), Chimaera (Gibbs et al. 2000), SiScan (Gibbs et al. 2000), MaxChi (Smith 1992), and LARD (Holmes et al. 1999). Recombination detected by at least three of the seven methods with a P value cutoff of 0.05 was considered as true recombination (Sabir et al. 2016). The recombinant sequences were removed and the procedure was repeated until no more recombination events were detected. We used SimPlot to detect interlineage recombination (Kolb et al. 2017). After removing interlineage recombination genome sequences, we used the GARD method implemented in the HyPhy software to detect intralineage recombination breakpoints on ORF1ab and the S gene (Pond et al. 2005; Dudas and Rambaut 2016). To visualize interlineage recombination events, alignments of the full genome were generated, and SplitsTree 5 was used to infer split decomposition networks (Huson and Bryant 2006).
Phylogenetic and Evolutionary Dynamic Analysis
Following recombination analysis, genomic sequences were separated at the breakpoint nucleotide positions 4989, 13680, and 22826. ML approaches were applied to the four segments and S gene coding region. ML trees were reconstructed using the general time-reversible substitution model while accounting for among-site heterogeneity through a discretized gamma distribution with four rate categories (GTR + Γ), and 1,000 bootstrap replicates were estimated using RAxML (version 8.4.10) (Stamatakis 2014). We used the same procedure to reconstruct an ML tree based on the nonrecombinant S gene sequences with three partitions for the codon positions. The same S gene sequences were used for phylodynamic analysis. We first assessed temporal signal in our data sets using TempEst (Rambaut et al. 2016) and continued to assess the best fitting model through marginal likelihood estimation using path-sampling and stepping-stone estimation approaches. This identified the relaxed molecular clock with an underlying lognormal distribution as the best fitting clock model (Baele et al. 2012, 2013). The tMRCA and evolutionary rate were estimated using the BEAST package (version 1.10.5) (Suchard et al. 2018), with separate GTR + Γ nucleotide substitution models on the three partitions based on the codon positions and an uncorrelated lognormal relaxed molecular clock (Drummond et al. 2006). In addition, we used a coalescent-based nonparametric skygrid prior for the tree topologies to model the effective population size over time (Gill et al. 2013). Two independent chains with a chain length of 1 × 108 converged to indistinguishable posterior distributions. Convergence and mixing were examined using the Tracer software (v1.7) (Rambaut et al. 2018) with a burn-in period of 10% of the total chain length. All parameter estimates yielded effective sampling size >200. The final MCC tree was generated by TreeAnnotator (version 1.10.5) and illustrated in Figtree (version 1.4.3) (http://tree.bio.ed.ac.uk/software/figtree/).
Discrete and Continuous Phylogeographic Analyses of PDCoV Spread in China
Discrete and continuous phylogeographic analyses of PDCoV China lineages were performed using BEAST 1.10.5 (Suchard et al. 2018) and the BEAGLE 3 library (Ayres et al. 2019) to improve computational performance. For both types of phylogeographic analyses, we specified a flexible substitution process modeled according to a GTR + Γ parametrization, a relaxed clock model with rates drawn from an underlying lognormal distribution (Drummond et al. 2006), and a flexible nonparametric skygrid coalescent model as the tree topology prior (Gill et al. 2013). The discrete phylogeographic analysis was performed using BSSVS to identify the best-supported among-location movement rates (Lemey et al. 2009) . In addition, the GLM extension of the discrete phylogeographic model (Lemey et al. 2014) was used to jointly infer the dispersal history of lineages among locations as well as the relevance and contribution of potential predictors to the transition rates between discrete locations. In practice, this approach allows to estimate the size of the contribution (expressed by the GLM coefficient) and the statistical support (expressed by a Bayes factor) for each potential predictor included in the model. Here, three different predictors were tested: 1) the geographic distance between sampling locations, as well as the average number of estimated annual live hogs in infected farms 2) at the locations of origin and 3) at the locations of destination. Discrete phylogeographic analyses (BSSVS and GLM analyses) were only based on the new PDCoV sequences presented in this study, that is, the sequences for which we had sampling locations allowing to integrate location-specific predictor values in the GLM analysis.
The continuous phylogeographic analysis was performed using two distinct models: the RRW diffusion model (Lemey et al. 2010) and the DRW diffusion model (Gill et al. 2017). In the RRW model, we used a lognormal distribution to model the among-branch heterogeneity in diffusion velocity. The DRW model combines the branch-specific diffusion rate variation with a constant directional trend (for movement along the latitude and longitude dimension) to preserve model identifiability (Gill et al. 2017). We use DRW to explore if the continuous phylogeographic reconstruction could be associated with a significant latitudinal and/or longitudinal drift. We performed two different analyses with the DRW model: a first analysis allowing both longitudinal and latitudinal drift, and because the resulting posterior estimates only provided evidence for a latitudinal drift, a second analysis only allowing a latitudinal drift. Contrary to the discrete phylogeographic analyses, the continuous analyses were based on both the new and the previously available PDCoV sequences from China (see supplementary table S2, Supplementary Material online). Because the sampling locations of several available sequences were only known at the province level, we used a sampling prior approach to define a potential area of origin for these sequences (Nylinder et al. 2014; Dellicour et al. 2020). The length of each BEAST inference procedure was adjusted according to the type of analysis: 300 million iterations for the discrete phylogeographic analyses and 500 million iterations for the continuous phylogeographic analyses. Parameter estimates were sampled every 100,000 generations, and the first 10% of the samples were removed as burn-in. Convergence and mixing properties were inspected using Tracer 1.7 (Rambaut et al. 2018) to ensure that effective sampling size values associated with relevant parameters were all >200. For the continuous phylogeographic reconstruction, MCC trees were obtained with TreeAnnotator 1.10.5 (Suchard et al. 2018) and based on 1,000 trees regularly sampled from each posterior distribution. For the visualization of the phylogeographic reconstruction performed with the DRW model, the MCC search was only based on posterior trees for which the root was positioned inland (i.e., not in the sea). We used R functions and scripts available in the package “seraphim” (Dellicour et al. 2016a, 2016b) to extract the spatiotemporal information embedded within the same 1,000 posterior trees and visualize the continuous phylogeographic reconstructions. We further used “seraphim” to estimate weighted lineage dispersal velocities from continuous phylogeographic reconstructions.
Selection and Protein Function Analysis
To detect selection on the S gene, an ML tree based on nonrecombinant sequences was reconstructed using Datamonkey (http://www.datamonkey.org/). The methods used to investigate positive amino acids sites included Single Likelihood Ancestor Counting (SLAC), Fixed Effects Likelihood (FEL), Mixed Effects Model of Evolution (MEME), and Fast Unconstrained Bayesian AppRoximation (FUBAR) for inferring selection (Kosakovsky Pond and Frost 2005; Murrell et al. 2012, 2013; Smith et al. 2015). The branch site REL and the GA-branch site models were chosen to detect selection pressure on individual branches. Codons were considered to be under selection if they were highlighted by at least three methods. Sites identified by at least two algorithms were considered as conservative positive selection. Protein models were created with PyMol (Molecular Graphics System, Version 2.0 Schrödinger, LLC, https://pymol.org/2/) using pdb file 6B7N representing the Cryo-EM structure of the S protein of the PDCoV (Shang et al. 2018). The distance measurement was done with the wizard tool of the program.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Professor Jiyong Zhou, Zhejiang University, and Professor Changchun Tu, Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, and Professor Weifeng Shi, Shandong First Medical University and Shandong Academy of Medical Sciences, for them guidance and help. We thank Haitao Qi of New Hope Liuhe Company and Ao Liao of Han Shiwei Company for providing pig samples. S.S., W.T.H., W.H., S.W., G.L., and L.Z. are financially supported by the National Natural Science Foundation of Outstanding Youth Fund in China (NSFC Grant No. 31922081), the National Key Research and Development Program of China (Grant No. 2016YFD0500402), The Young Top-Notch Talents of National Ten Thousand Talent Program, the Fundamental Research Funds for the Central Universities (Grant No. Y0201900459), Six Talent Peaks Project of Jiangsu Province of China (Grant No. NY-045), Sino German Cooperation and Exchange Project in International Cooperation and Cultivation Project in 2019, and the Bioinformatics Center of Nanjing Agricultural University. S.D. and M.G. are supported by the Fonds National de la Recherche Scientifique (FRS-FNRS, Belgium). X.J. and M.A.S. are partially supported through National Institutes of Health (Grant No. U19 AI135995). G.B. acknowledges support from the Interne Fondsen KU Leuven/Internal Funds KU Leuven (Grant No. C14/18/094) and the Research Foundation—Flanders (“Fonds voor Wetenschappelijk Onderzoek—Vlaanderen,” G0E1420N). P.L. acknowledges support by the European Research Council under the European Union’s Horizon 2020 research and innovation program (Grant No. 725422-ReservoirDOCS) and the Research Foundation—Flanders (“Fonds voor Wetenschappelijk Onderzoek—Vlaanderen,” G066215N, G0D5117N, and 75 G0B9317N).
References
- Ayres DL, Cummings MP, Baele G, Darling AE, Lewis PO, Swofford DL, Huelsenbeck JP, Lemey P, Rambaut A, Suchard MA.. 2019. BEAGLE 3: improved performance, scaling, and usability for a high-performance computing library for statistical phylogenetics. Syst Biol. 68(6):1052–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV.. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 29(9):2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, Li WL, Drummond AJ, Suchard MA, Lemey P.. 2013. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol. 30(2):239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni MF, Posada D, Feldman MW.. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176(2):1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong R, Shi M, Grueber CE, Holmes EC, Hogg CJ, Belov K, Barrs VR.. 2019. Fecal viral diversity of captive and wild Tasmanian devils characterized using virion-enriched metagenomics and metatranscriptomics. J Virol. 93(11):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Baldwin HJ, Tateno AF, Zerbinati RM, Annan A, Owusu M, Nkrumah EE, Maganga GD, Oppong S, Adu-Sarkodie Y, et al. 2015. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol. 89(23):11858–11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Eckerle I, Memish ZA, Liljander AM, Dijkman R, Jonsdottir H, Juma Ngeiywa KJ, Kamau E, Younan M, Al Masri M, et al. 2016. Link of a ubiquitous human coronavirus to dromedary camels. Proc Natl Acad Sci U S A. 113(35):9864–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellicour S, Rose R, Pybus OG. 2016. a. Explaining the geographic spread of emerging epidemics: a framework for comparing viral phylogenies and environmental landscape data. BMC Bioinformatics 17(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellicour S, Rose R, Faria NR, Lemey P, Pybus OG. 2016. b. SERAPHIM: studying environmental rasters and phylogenetically informed movements. Bioinformatics 32(20):3204–3206. [DOI] [PubMed] [Google Scholar]
- Dellicour S, Lemey P, Artois J, Lam TT, Fusaro A, Monne I, Cattoli G, Kuznetsov D, Xenarios I, Dauphin G, et al. 2020. Incorporating heterogeneous sampling probabilities in continuous phylogeographic inference—application to H5N1 spread in the Mekong region. Bioinformatics 36(7):2098–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellicour S, Rose R, Faria NR, Vieira LFP, Bourhy H, Gilbert M, Lemey P, Pybus OG.. 2017. Using viral gene sequences to compare and explain the heterogeneous spatial dynamics of virus epidemics. Mol Biol Evol. 34(10):2563–2571. [DOI] [PubMed] [Google Scholar]
- Dellicour S, Troupin C, Jahanbakhsh F, Salama A, Massoudi S, Moghaddam MK, Baele G, Lemey P, Gholami A, Bourhy H.. 2019. Using phylogeographic approaches to analyse the dispersal history, velocity and direction of viral lineages—application to rabies virus spread in Iran. Mol Ecol. 28(18):4335–4350. [DOI] [PubMed] [Google Scholar]
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RAM, et al. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 348(20):1967–1976. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A.. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4(5):e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas G, Rambaut A.. 2016. MERS-CoV recombination: implications about the reservoir and potential for adaptation. Virus Evol. 2(1):vev023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D, Cagliani R, Clerici M, Sironi M.. 2017. Molecular evolution of human coronavirus genomes. Trends Microbiol. 25(1):35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs MJ, Armstrong JS, Gibbs AJ.. 2000. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16(7):573–582. [DOI] [PubMed] [Google Scholar]
- Gilbert M, Conchedda G, Van Boeckel TP, Cinardi G, Linard C, Nicolas G, Thanapongtharm W, D’Aietti L, Wint W, Newman SH, et al. 2015. Income disparities and the global distribution of intensively farmed chicken and pigs. PLoS One 10(7):e0133381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MS, Tung Ho LS, Baele G, Lemey P, Suchard MA. 2017. A Relaxed Directional Random Walk Model for Phylogenetic Trait Evolution. Syst Biol. 66(3):299–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA.. 2013. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol. 30(3):713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Fang L, Ye X, Chen J, Xu S, Zhu X, Miao Y, Wang D, Xiao S.. 2019. Evolutionary and genotypic analyses of global porcine epidemic diarrhea virus strains. Transbound Emerg Dis. 66(1):111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Auclert LZ, Zhai X, Wong G, Zhang C, Zhu H, Xing G, Wang S, He W, Li K, et al. 2019. Interspecies transmission, genetic diversity, and evolutionary dynamics of pseudorabies virus. J Infect Dis. 219(11):1705–1715. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Worobey M, Rambaut A.. 1999. Phylogenetic evidence for recombination in dengue virus. Mol Biol Evol. 16(3):405–409. [DOI] [PubMed] [Google Scholar]
- Hu H, Jung K, Vlasova AN, Chepngeno J, Lu Z, Wang Q, Saif LJ.. 2015. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J Clin Microbiol. 53(5):1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YW, Dickerman AW, Pineyro P, Li L, Fang L, Kiehne R, Opriessnig T, Meng XJ.. 2013. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio 4(5):e00737–00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23(2):254–267. [DOI] [PubMed] [Google Scholar]
- Jung K, Hu H, Eyerly B, Lu Z, Chepngeno J, Saif LJ.. 2015. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg Infect Dis. 21(4):650–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb AW, Lewin AC, Moeller Trane R, McLellan GJ, Brandt CR.. 2017. Phylogenetic and recombination analysis of the herpesvirus genus Varicellovirus. BMC Genomics. 18(1):887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD.. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 22(5):1208–1222. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Lee P, Tsang AK, Yip CC, Tse H, Lee RA, So LY, Lau YL, Chan KH, Woo PC, et al. 2011. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol. 85(21):11325–11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SKP, Wong EYM, Tsang CC, Ahmed SS, Au-Yeung RKH, Yuen KY, Wernery U, Woo P.. 2018. Discovery and sequence analysis of four deltacoronaviruses from birds in the Middle East reveal interspecies jumping with recombination as a potential mechanism for avian-to-avian and avian-to-mammalian transmission. J Virol. 92(15):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le VP, Song S, An BH, Park GN, Pham NT, Le DQ, Nguyen VT, Vu TTH, Kim KS, Choe S, et al. 2018. A novel strain of porcine deltacoronavirus in Vietnam. Arch Virol. 163(1):203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Chung HC, Nguyen VG, Moon HJ, Kim HK, Park SJ, Lee CH, Lee GE, Park BK.. 2016. Detection and phylogenetic analysis of porcine deltacoronavirus in Korean swine farms, 2015. Transbound Emerg Dis. 63(3):248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee C.. 2014. Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2(6):e01191–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Welch JJ, Suchard MA. 2010. Phylogeography takes a relaxed random walk in continuous space and time. Mol Biol Evol. 27(8):1877–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P Rambaut A Bedford T Faria N Bielejec F Baele G Russell CA Smith DJ Pybus OG Brockmann D, . et al. 2014. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 10(2):e1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol. 5(9):e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Feng H, Liu Y, Chen Y, Wei Q, Wang J, Liu D, Huang H, Su Y, Wang D, et al. 2018. Molecular evolution of porcine epidemic diarrhea virus and porcine deltacoronavirus strains in Central China. Res Vet Sci. 120:63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Chen Q, Harmon KM, Yoon KJ, Schwartz KJ, Hoogland MJ, Gauger PC, Main RG, Zhang J.. 2014. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2(2):e00278–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, He W, Zhu H, Bi Y, Wang R, Xing G, Zhang C, Zhou J, Yuen KY, Gao GF, et al. 2018. Origin, genetic diversity, and evolutionary dynamics of novel porcine circovirus 3. Adv Sci (Weinh). 5(9):1800275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch BJ.. 2018. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci U S A. 115(22):E5135–E5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhang X, Zhang Z, Chen R, Zhang Z, Xue Q.. 2017. Complete genome characterization of novel Chinese porcine deltacoronavirus strain SD. Genome Announc. 5(40):e00930–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsirigool A, Saeng-Chuto K, Madapong A, Temeeyasen G, Tripipat T, Kaewprommal P, Tantituvanont A, Piriyapongsa J, Nilubol D.. 2017. The genetic diversity and complete genome analysis of two novel porcine deltacoronavirus isolates in Thailand in 2015. Virus Genes 53(2):240–248. [DOI] [PubMed] [Google Scholar]
- Lorsirigool A, Saeng-Chuto K, Temeeyasen G, Madapong A, Tripipat T, Wegner M, Tuntituvanont A, Intrakamhaeng M, Nilubol D.. 2016. The first detection and full-length genome sequence of porcine deltacoronavirus isolated in Lao PDR. Arch Virol. 161(10):2909–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J.. 2015. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio 6(2):e00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madapong A, Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Srijangwad A, Tripipat T, Wegner M, Nilubol D.. 2016. Complete genome sequence of porcine deltacoronavirus isolated in Thailand in 2015. Genome Announc. 4(3):e00408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Rybicki E.. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16(6):562–563. [DOI] [PubMed] [Google Scholar]
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B.. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 1(1):vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Pond SLK, Scheffler K.. 2013. FUBAR: a Fast, Unconstrained Bayesian AppRoximation for inferring selection. Mol Biol Evol. 30(5):1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Pond S.. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8(7):e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylinder S, Lemey P, De Bruyn M, Suchard MA, Pfeil BE, Walsh N, Anderberg AA.. 2014. On the biogeography of Centipeda: a species-tree diffusion approach. Syst Biol. 63(2):178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padidam M, Sawyer S, Fauquet CM.. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265(2):218–225. [DOI] [PubMed] [Google Scholar]
- Perez-Rivera C, Ramirez-Mendoza H, Mendoza-Elvira S, Segura-Velazquez R, Sanchez-Betancourt JI.. 2019. First report and phylogenetic analysis of porcine deltacoronavirus in Mexico. Transbound Emerg Dis. 66:1436–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV.. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21(5):676–679. [DOI] [PubMed] [Google Scholar]
- Pybus OG, Suchard MA, Lemey P, Bernardin FJ, Rambaut A, Crawford FW, Gray RR, Arinaminpathy N, Stramer SL, Busch MP, et al. 2012. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc Natl Acad Sci U S A. 109(37):15066–15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Dong X, Baele G, Suchard MA.. 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol. 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Lam TT, Max Carvalho L, Pybus OG.. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2(1):vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir JS, Lam TT, Ahmed MM, Li L, Shen Y, Abo-Aba SE, Qureshi MI, Abu-Zeid M, Zhang Y, Khiyami MA, et al. 2016. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 351(6268):81–84. [DOI] [PubMed] [Google Scholar]
- Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Vui DT, Stott CJ, Madapong A, Tripipat T, Wegner M, Intrakamhaeng M, Chongcharoen W, et al. 2017. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound Emerg Dis. 64(1):3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Zheng Y, Yang Y, Liu C, Geng Q, Tai W, Du L, Zhou Y, Zhang W, Li F.. 2018. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J Virol. 92(4):e01556–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459(7250):1122–1125. [DOI] [PubMed] [Google Scholar]
- Smith JM. 1992. Analyzing the mosaic structure of genes. J Mol Evol. 34(2):126–129. [DOI] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Pond S.. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 32(5):1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF.. 2016. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 24(6):490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A.. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4(1):vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, He W-T, Wang L, Lai A, Ji X, Zhai X, Li G, Suchard MA, Tian J, Zhou J, et al. 2020. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med. 26(5):483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MH, Deng MC, Chung YH, Huang YL, Chang CY, Lan YC, Chou HL, Chao DY.. 2015. Evolutionary characterization of the emerging porcine epidemic diarrhea virus worldwide and 2014 epidemic in Taiwan. Infect Genet Evol. 36:108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Shi M, Chommanard C, Queen K, Zhang J, Markotter W, Kuzmin IV, Holmes EC, Tong S.. 2017. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol. 91(5):e01953–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Feng Y, Vrancken B, Cazelles B, Tan H, Gill MS, Yang Q, Li Y, Yang W, Zhang Y, et al. 2018. Transmission dynamics of re-emerging rabies in domestic dogs of rural China. PLoS Pathog. 14(12):e1007392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Liu Y, Ji CM, Yang YL, Liang QZ, Zhao P, Xu LD, Lei XM, Luo WT, Qin P, et al. 2018. Porcine deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J Virol. 92(12):e00318–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Byrum B, Zhang Y.. 2014. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis. 20(7):1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, et al. 2012. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 86(7):3995–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Yip CC, Huang Y, Tsoi HW, Chan KH, Yuen KY.. 2006. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol. 80(14):7136–7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Tortorici MA, Snijder J, Yoshioka C, Walls AC, Li W, McGuire AT, Rey FA, Bosch BJ, Veesler D.. 2018. Glycan shield and fusion activation of a deltacoronavirus spike glycoprotein fine-tuned for enteric infections. J Virol. 92:e01628–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA.. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 367(19):1814–1820. [DOI] [PubMed] [Google Scholar]
- Zhang MJ, Liu DJ, Liu XL, Ge XY, Jongkaewwattana A, He QG, Luo R.. 2019. Genomic characterization and pathogenicity of porcine deltacoronavirus strain CHN-HG-2017 from China. Arch Virol. 164(2):413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng Y, Xing G, Yu J, Liao A, Du L, Lei J, Lian X, Zhou J, Gu J.. 2019. Detection and spike gene characterization in porcine deltacoronavirus in China during 2016–2018. Infect Genet Evol. 73:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Fan H, Lan T, Yang X-L, Shi W-F, Zhang W, Zhu Y, Zhang Y-W, Xie Q-M, Mani S, et al. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556(7700):255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XY, Wang D, Zhou JW, Pan T, Chen JY, Yang YT, Lv MT, Ye X, Peng GQ, Fang LR, et al. 2017. Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J Virol. 91(10):e00003–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.