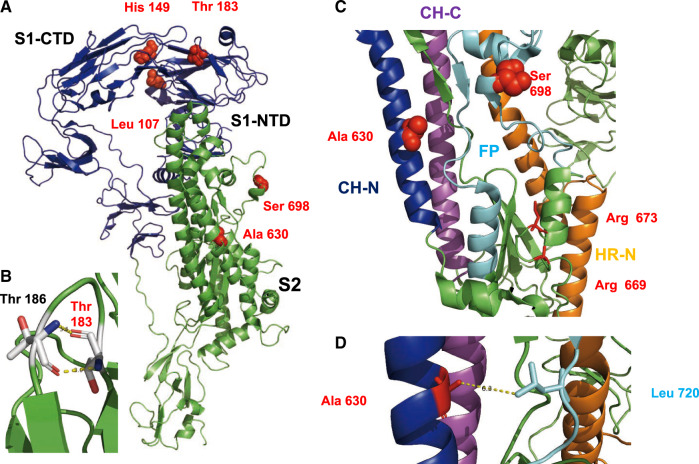
Fig. 5.
Location of selected amino acids in the structure of S protein. (A) Cartoon representation of an S monomer. The S1 subunit is represented in blue and the S2 subunit in green. Selected amino acids are shown as red spheres. Leu 107 and His 149 are located in the N-terminal domain of S1 (S1-NTD) that binds to unidentified sugars, but close to the C-terminal domain of S1 (S1-CTD) that contains the binding site for the protein receptor. (B) Hydrogen bonds formed between Thr 183 and Thr 186 that might stabilize a loop. (C) Part of the structure of S2 with individual elements drawn in different colors. Central helix N (CH-N) is represented in blue, the central helix C (CH-C) in magenta, the fusion peptide (FP) in cyan, and the heptad repeat N in orange. Arg 669 and Arg 673, shown as red sticks, are presumed proteolytic cleavage sites. The selected amino acids Ala 630 and Ser 698 are shown as red spheres. (D) Detail of S2 showing the selected amino acid Ala 630 in proximity (6.3 Å) to Leu 720 in the fusion peptide.