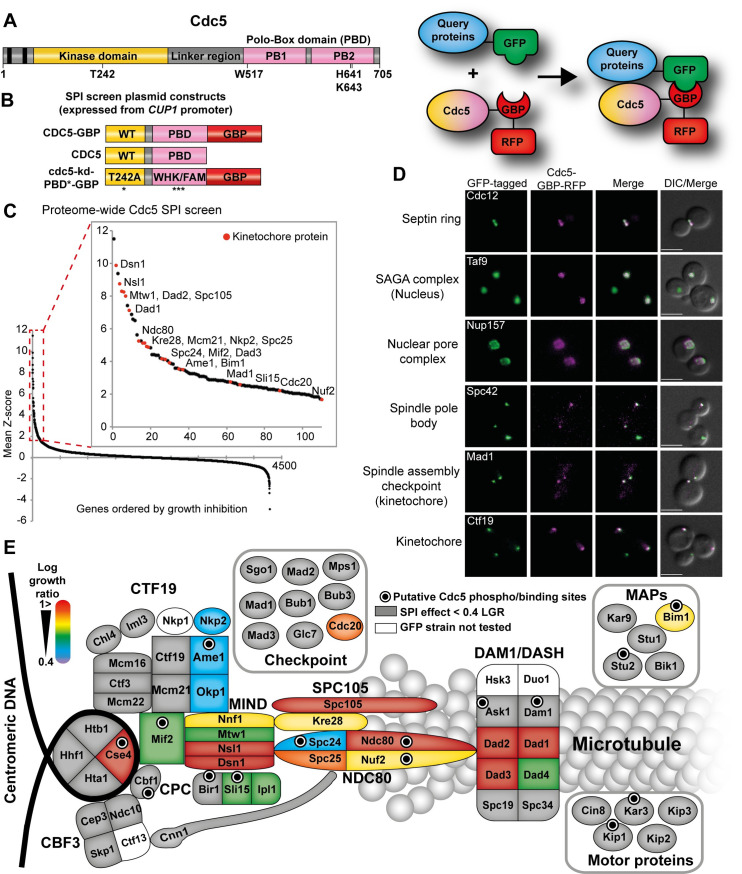
Fig 2. Proteome-wide Cdc5 SPI screen is enriched for kinetochore proteins.
(A) The key domains of Polo kinase Cdc5 are shown. The N-terminal kinase domain contains a threonine 242 residue when substituted to alanine creates a catalytic-inactive mutant. The C-terminal non-catalytic domain contains two polo-boxes (PB1 and PB2), together called the polo-box domain (PBD) which binds to previously phosphorylated sites which targets the kinase domain of Cdc5 to its substrates and facilitates the phosphorylation at another site on the same substrate or surrounding substrates. A flexible linker region joins the kinase domain and PBD. (B) Schematic of the Cdc5-GBP constructs and controls used in the Cdc5 SPI assays (left). Expression of all constructs are controlled by CUP1 promoter. The cdc5-kd-PBD*-GBP control contains a mutated form of PBD (W517F, H641A, K643M). The GBP includes an RFP tag. The inset on the right shows a cartoon displaying the Cdc5-GBP interaction with a query GFP-tagged protein. (C) Data from the proteome-wide Cdc5 SPI screen are plotted as in Fig 1A. The inset shows that many kinetochore proteins (red dots) are among the top 100 Cdc5 SPIs. (D) Live imaging of cells containing different GFP-tagged proteins and expressing Cdc5-GBP shows that Cdc5-GBP can be constitutively recruited to many different subcellular locations as judged by colocalization. Scale bars are 5μm. (E) An illustration of the Cdc5 kinetochore SPIs, mapped onto a cartoon representation of the kinetochore. The color-coded map is based on log growth ratios of Cdc5-GBP compared with the average of both kinase-dead controls. The strength of the growth inhibition caused by Cdc5 association is indicated by a color-coded scale with high log growth ratios (LGR > 1) shown in red. Forced interactions that do not produce a growth phenotype are shown in grey (LGR < 0.4). Proteins that contain phosphosites that fit Cdc5 consensus for either phosphorylation or binding (E/D/Q)-X-(pS/T)-X and S-pS/T-P, respectively) are indicated with black dots.