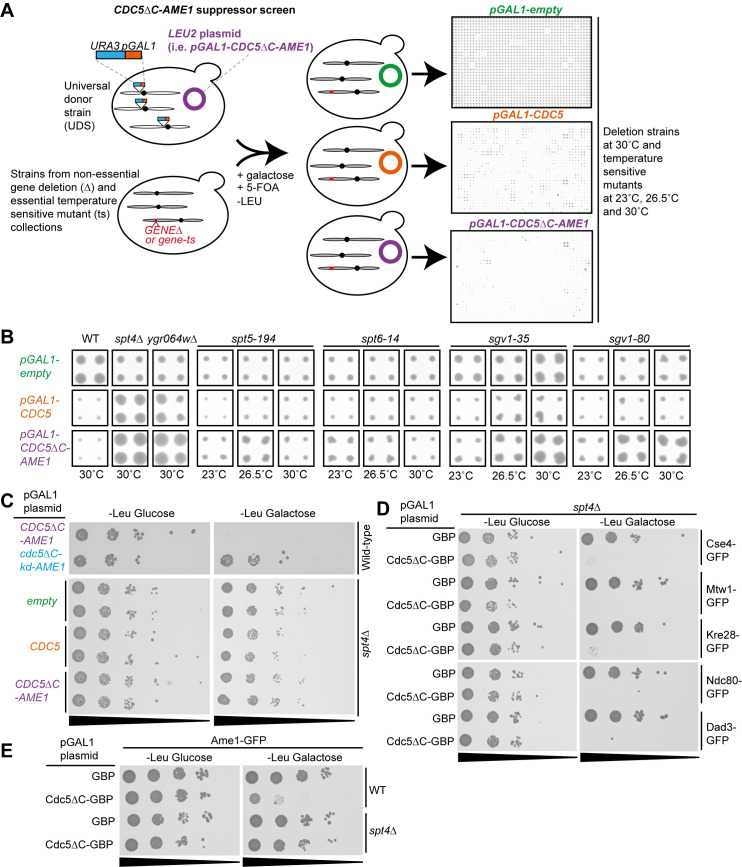
Fig 5. Suppressors of the forced Cdc5-Ame1 interaction phenotype.
(A) Diagram showing the experimental layout of the suppressor screen. Selective ploidy ablation (SPA) was used to introduce pGAL1-CDC5ΔC-AME1, pGAL1-CDC5 and pGAL1-empty plasmids into the deletion (Δ) and temperature-sensitive mutant (ts) collections. The Δ screen was performed at 30°C and the ts screen at 23°C, 26.5°C and 30°C. The agar plates from the final selection step were scanned after three days and representative examples are shown (see methods for further details). (B) A selection of cropped images of colonies from the suppressor screens showing that growth defects caused by pGAL1-CDC5ΔC-AME1 or pGAL1-CDC5 are suppressed by genetic deletions or mutations. (C) 10-fold serial dilutions spot assay with wild-type and spt4Δ strains containing pGAL1-CDC5ΔC-AME1 and control plasmids shows that spt4Δ suppresses growth defect caused by CDC5ΔC-AME1 expression. (D) 10-fold serial dilutions spot assay with GFP-tagged kinetochore strains (Cse4-GFP (internally tagged), Mtw1-GFP, Kre28-GFP, Ndc80-GFP and Dad3-GFP) containing pGAL1-CDC5ΔC-GBP and pGAL1-GBP plasmids shows that spt4Δ is not sufficient to suppress the growth defect caused by constitutive localization of Cdc5 to these proteins. (E) 10-fold serial dilutions spot assay with wild-type and spt4Δ strains encoding Ame1-GFP and pGAL1-CDC5ΔC-GBP and pGAL1-GBP plasmids showing that spt4Δ suppressed growth defect caused by constitutive Cdc5 association with Ame1.