Extended Data Figure 2. Further details of effects of SEPHS2 KO and overexpression in cells and in the MDA231 orthotopic xenograft.
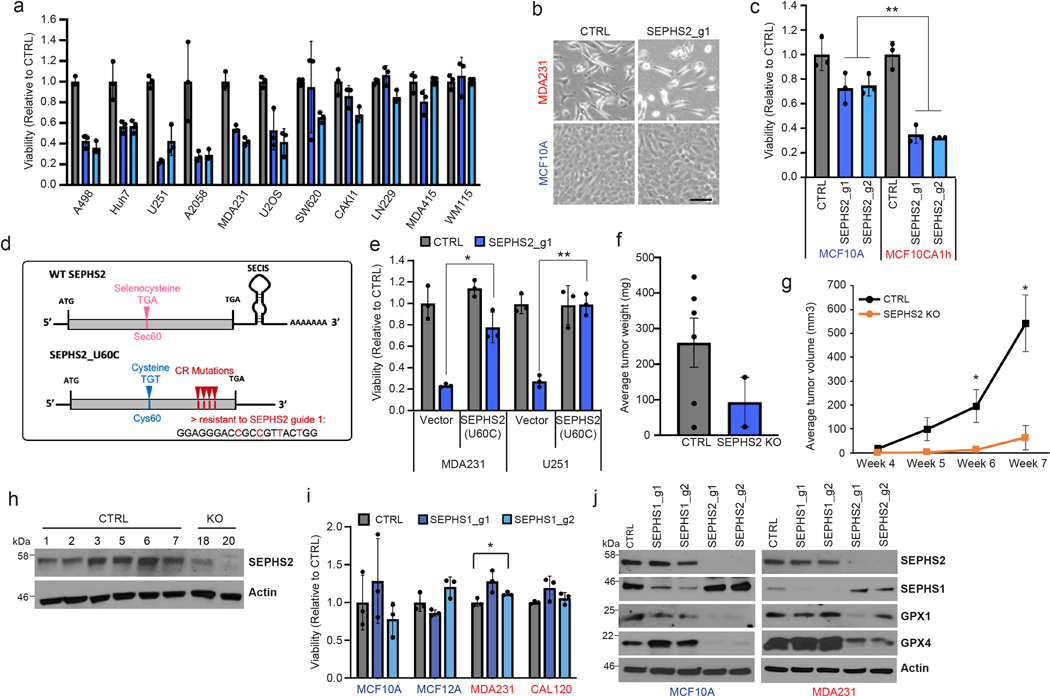
(a) Viability of cell lines following KO with guides against SEPHS2 for 9 days. Values are relative to the same cell lines expressing nontargeting (CTRL) guide (=1.0), (b) Light microscope images of MDAMB231 and MCF10A cells subjected to CTRL or SEPSECS KO. Scale bar=25μm. (c) Viability of cell lines following KO with guides against SEPHS2 for 11 days. Values are relative to the same cell lines expressing nontargeting guide (=1.0). (d) Map of wild type SEPHS2 and CRISPR resistant mutant SEPHS2_U60C. Details of features are provided in Methods. (e) Viability of vector and SEPHS2_U60C overexpressing cell lines subjected to CTRL and SEPHS2 g1 KO. (f) Weight measurements for ex vivo CTRL and SEPHS2 KO MDAMB231 orthotopic xenograft tumors. (g) In vivo tumor volume measurements for CTRL and SEPHS2 KO MDAMB231 orthotopic xenograft tumors. For f and g, if no tumor has formed, tumor weight and volume was designated 0 for CTRL, n=6 and for SEPHS2 KO n=2. (h) Immunoblot of SEPHS2 in CTRL and SEPHS2 KO MDAMB231 orthotopic xenograft tumors. Number is an animal identifier number. For a,c,e, n=3 biological replicates; error bars are S.D. Number is an animal identifier number. (i) Viabilities of noncancer (immortalized) and cancer lines following KO with guides against SEPHS1 (blue bars) for 9 days. Values are relative to the same cell lines expressing nontargeting guide (black bars), set at 1.0. (j) Immunoblots of selenoprotein expression in MCF10A and MDAMB231 cells subjected to (CRISPR/Cas9-induced) KO with guides against SEPHS1, SEPHS2, or control guides for 11 days. For a,c,e and i, n=3 biological replicates; error bars are S.D. For all panels, the measure of center is mean. *p>0.05, **p<0.01 (student’s two-tailed t test).
