Abstract
Allergic disorders, characterized by Th2 immune responses to environmental substances, are increasingly common in children in Western societies. Multiple studies indicate that breastfeeding, early complementary introduction of food allergens, and antibiotic avoidance in the first year of life reduces allergic outcomes in at-risk children. Why the benefit of these practices is restricted to early life is largely unknown. We identified a preweaning interval during which dietary antigens are assimilated by the colonic immune system. This interval is under maternal control via temporal changes in breast milk, coincides with an influx of naive T cells into the colon, and is followed by the development of a long-lived population of colonic peripherally derived Tregs (pTregs) that can be specific for dietary antigens encountered during this interval. Desynchronization of mothers and offspring produced durable deficits in these pTregs, impaired tolerance to dietary antigens introduced during and after this preweaning interval, and resulted in spontaneous Th2 responses. These effects could be rescued by pTregs from the periweaning colon or by Tregs generated in vitro using periweaning colonic antigen-presenting cells. These findings demonstrate that mothers and their offspring are synchronized for the development of a balanced immune system.
Keywords: Gastroenterology, Immunology
Keywords: Allergy, Tolerance
Mothers and offspring are synchronized through chronologic changes in breastmilk to promote tolerance to dietary antigens and restrain Th2 responses
Introduction
Allergic diseases are prevalent, complex, and heterogenous disorders that are rapidly increasing in Westernized societies (1–3). These disorders are characterized by Th2 responses to environmental antigens, and risk factors for the development of allergy include polymorphisms in genes related to the generation of Th2 responses (4). However, changes in inherited factors alone are insufficient to explain this rapid increase. Individuals emigrating from countries with a low prevalence of allergies to one with a high prevalence of allergy have an increased frequency of allergic disorders, which was even further increased in the following generation (5). This observation is consistent with early life environmental influences encountered by the second generation, which was born in a region with a high prevalence of allergy, as contributors to the rapid rise in allergic diseases. However, the contribution of environmental factors to the rise in allergic diseases is complex, as environmental exposures can also reduce allergy.
Decreased exposure to microbes is associated with increased allergic outcomes, suggesting that the lack of exposure to microbes, or the “hygiene hypothesis,” is a risk for developing allergy (6–8). Supporting this concept, germ-free mice have exaggerated Th2 responses (9, 10), providing a causal link between the lack of exposure to microbes and allergic responses. Beyond the general loss of exposure to microbes, alterations in the gut microbiota, or dysbiosis, are implicated in allergic disease. Dysbiosis of the gut microbiota has been observed in individuals with allergy, and transfer of the dysbiotic gut microbiota from individuals with allergy to mice potentiates an allergic phenotype (11). Moreover, specific taxa of the gut microbiota can protect against an allergic phenotype in mice (11, 12). Together, these observations indicate that loss of exposure to some gut bacteria may be an environmental risk factor for the development of allergy. This effect of gut bacteria to reduce allergy may be through the induction of peripherally induced Tregs (pTregs), as the loss of pTregs or pTreg dysfunction is associated with exaggerated Th2 responses and allergy (13–17), and specific gut bacterial taxa with the property to potently induce pTregs have been identified in mice and humans (18, 19).
However, the relationship between beneficial environmental exposures and the reduction of allergic outcomes may not be straightforward. The effect of disruption of the gut microbiota on allergic outcomes is more pronounced early in life, when these tolerance-inducing taxa are less abundant. Indeed, the use of antibiotics in children in the first year of life has a strong association with increased allergic outcomes (20–22), and children at risk for developing asthma later in life have a transient dysbiosis of the gut microbiota in the first few months of life (23). Furthermore, studies in mice reveal that antibiotic therapy given before weaning, but not later, results in allergic outcomes (24, 25) and that colonization of germ-free mice before weaning, but not as adults, reduces allergic outcomes (26). Furthermore, disruption of the “weaning reaction,” an inflammatory reaction in the terminal ileum to gut microbes during a specific period around the time of weaning, correlated with an increased propensity for immune pathologies, which correlated with the loss of a population pTregs (27). In aggregate, these observations support that some benefits of the exposure to specific gut microbes have a time-dependent nature that is restricted to a period in early life.
There is also a time-dependent nature to the benefits of exposure to food allergens via the gastrointestinal tract to reduce food allergy. Oral tolerance is a well-recognized and well-studied phenomenon in which ingestion of a dietary antigen induces antigen-specific Tregs that suppress subsequent immune responses to that antigen (28). Oral tolerance induction is not restricted to one time in life. Studies in mice suggest that the pTregs induced by dietary antigens require repeated antigen exposure to persist and that these pTregs can suppress food allergy responses (29). However, observational studies and clinical trials have found a time-dependent nature to the benefit of food allergen introduction for the prevention of food allergy in children. Introduction of food allergens before or around 6 months of life, but not later, reduces subsequent food allergy (30–33). Furthermore, this benefit is most advantageous for breastfed children (34, 35) and is durable even in the absence of continual exposure to the food allergen (36). Thus, while environmental exposures reducing allergic outcomes have been identified, many facets of how these exposures mediate the observed benefits and what restricts the benefits of these exposures to a time in early life remain largely unknown.
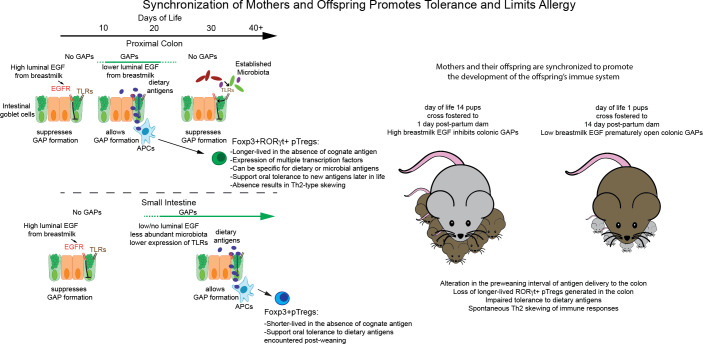
Here, we identify a preweaning interval during which dietary antigens are delivered to the colonic immune system to generate a population of long-lived pTregs. The timing of this interval delivering dietary antigens to the colonic immune system is under maternal control through chronological changes in breast milk epidermal growth factor (EGF) as opposed to other potential intrinsic sources of gut lumen EGF such as salivary glands, Brunner’s glands, the pancreas, and Paneth cells (37–39). Desynchronizing dams and pups alters the delivery of luminal antigens to the immune system. This results in abrogation of tolerance to dietary antigens encountered during this interval, reduction of this pTreg population, and long-term effects, including Th2 skewing and impaired tolerance to newly encountered dietary antigens. These deficits can be restored by adoptive transfer of pTregs from the periweaning colon or by adoptive transfer of Tregs generated in vivo using antigen-presenting cells (APCs) from the preweaning colon, demonstrating that these Tregs are sufficient to restrain Th2 skewing and impaired tolerance seen following desynchronization of dams and pups. In total, these observations give insight into the biological basis for the benefits of breastfeeding and complementary introduction of food allergens on the reduction of allergic outcomes, and they indicate that mothers and their offspring are synchronized to promote tolerance to food allergens introduced before weaning and to promote the development of a balanced immune system.
Results
Chronological changes in breast milk regulate GAPs to control regional and temporal luminal antigen exposure in the offspring’s intestine.
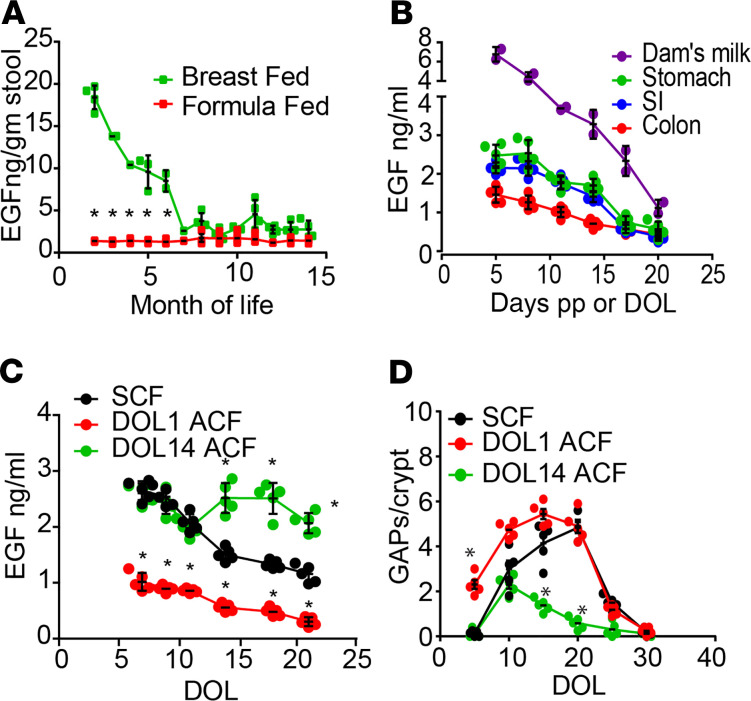
Previously, we identified a preweaning interval in mice, day of life 10 (DOL10) to around the time of weaning (DOL21), during which goblet cells in the cecum, ascending colon, transverse colon, and proximal descending colon, referred to as the proximal colon, form goblet cell–associated antigen passages (GAPs) and deliver commensal bacterial antigens to the colonic immune system to generate antigen-specific tolerance to some gut commensals (40). Whether this interval and this process are restricted to the assimilation of bacterial antigens, which are largely limited to the colon, or extends to dietary antigens, which are largely assimilated by the small intestine (SI) immune system in adults (29), and how the timing of this interval is controlled are unknown. Activation of the EGF receptor (EGFR) expressed by goblet cells suppresses the ability of goblet cells in the SI and proximal colon to respond to acetylcholine to form GAPs and accordingly suppresses the delivery of luminal antigens to lamina propria (LP) APCs in the SI and proximal colon (41–43). High levels of EGF in the gut lumen inhibit colonic GAP formation before DOL10, and after DOL10, luminal EGF levels fall to allow colonic GAPs to form to initiate this preweaning interval (40). However, the source of EGF and how EGF levels in the gut lumen are controlled to define this preweaning interval are unclear. EGF is highly expressed at multiple sites in the gastrointestinal tract, including salivary glands, Brunner’s glands in the duodenum, the pancreas, and Paneth cells, which are located throughout the SI (37–39). In addition to these sources of EGF, which are intrinsic to the offspring, EGF is also found in breast milk (44, 45), suggesting the potential for maternal influences on this process. To gain insight into the sources of EGF controlling these events, we evaluated EGF levels in stools from children who were exclusively breastfed or exclusively formula fed. Stools from breastfed children contained significantly higher levels of EGF that decreased over the first 6 months of life until weaning when compared with formula-fed children (Figure 1A), consistent with a maternal origin for EGF in the children’s gut. The temporal decrement in EGF levels in the stools of breastfed children could reflect a temporal decrement in breast milk EGF concentrations or, alternatively, might reflect changes occurring within the offspring’s developing gastrointestinal tract, such as increased production of digestive enzymes, increased absorptive capacity, increased intestinal secretion, and/or lengthening of the gut, which would decrease the luminal EGF concentrations. We evaluated the temporal concentration of EGF in breast milk of lactating mice and correlated this with EGF levels in the nursing pup’s gastrointestinal tract. We observed that the concentration of EGF in breast milk was greater than that seen in the pups’ GI tract and decreased over time (Figure 1B). Moreover, the nursing pups had temporal decrement in EGF corresponding to the temporal decrement of EGF seen in breast milk and a proximal to distal gradient of EGF in their GI tract, with highest levels seen in the stomach contents (Figure 1B). These observations implicate breast milk as a source of EGF in the nursing offspring’s GI tract and raise the possibility that the initiation of the preweaning interval during which proximal colonic GAPs are present could be under maternal control.
Figure 1. Maternal control of goblet cell–associated antigen passages in the preweaning period.
(A) EGF concentration in stools obtained from healthy formula-fed or healthy breastfed children through the first year of life. (B) EGF concentration in lactating dam’s milk, or the pups’ stomach, small intestine (SI), or colon contents, by postpartum (pp) days or day of life (DOL), respectively. (C and D) DOL1 pups were asynchronously cross-fostered to dams 14 day pp (DOL1 ACF), DOL14 pups were asynchronously cross-fostered to dams 1 day pp (DOL14 ACF), or DOL1 pups were synchronously cross-fostered to dams 1 day pp (SCF). EGF concentration in stomach contents (C), and density of GAPs per colonic crypt (D). Data are presented as the mean ± SEM, *P < 0.05. A represents data from 42 stool specimens from 6 breastfed children and 53 stool specimens from 8 formula-fed children; n = 2 dams and 5 pups per time point for B; and n = 5 mice per time point for C and D. Pups from 2 litters per condition and/or time point were used for B–D. Significance calculated using 2-tailed Student’s t test in A, and 2-way ANOVA with a Dunnett’s post hoc test in C and D.
In conventionally reared mice, we found that colonic GAPs begin to form around DOL10 and regress around the time of weaning (40) (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137943DS1). SI and colonic GAPs form independent of gut microbiota, as evidenced by the normal timing of SI and colonic GAP formation in germ-free mice (Supplemental Figure 1). In contrast to SPF housed mice, colonic GAPs persisted after weaning in germ-free mice (Supplemental Figure 1), which is consistent with the fully developed gut microbiota inhibiting colonic GAPs in adult mice (41). The gut microbiota and microbial products do not inhibit SI GAPs, in part due to lower expression of TLRs in SI goblet cells (41). To evaluate if luminal EGF concentrations and colonic GAP formation in the preweaning pups were indeed driven by the dam’s timing postpartum (pp), we cross-fostered pups to dams that delivered on the same day (synchronous cross-foster; SCF) and to dams that had delivered 2 weeks earlier or later (asynchronous cross-foster; ACF). We observed that DOL1 mice SCF to dams 1 day pp displayed a temporal pattern of EGF concentrations in the stomach contents and colonic GAP density similar to those of conventionally reared mice: nursing mice that had not been cross-fostered (Figure 1, C and D, compare with Figure 1B and Supplemental Figure 1). However, DOL1 ACF mice exposed to dams 2 weeks pp had significantly decreased EGF concentrations in the stomach contents during the preweaning interval and significantly increased colonic GAP density before DOL10, when GAPs normally appear in the colon (Figure 1, C and D, and Supplemental Figure 1). The density of GAP formation is also dependent on goblet cell density, which increases throughout the preweaning period. DOL1 ACF mice and SCF mice displayed similar kinetics and density of colonic GAPs in the peri- and postweaning period (Figure 1D), where GAP density is largely regulated by goblet cell sensing of the gut microbiota, which inhibits GAP formation when low EGF levels allow GAPs to form. Conversely, DOL14 ACF mice exposed to dams 1 day pp demonstrated significantly increased luminal EGF concentrations following ACF and significantly decreased colonic GAPs following ACF up to approximately DOL25, when GAPs normally become rare due to inhibition by the gut microbiota (Figure 1, C and D). These data indicate that the dam’s timing pp controls a pathway that can deliver luminal substances to the preweaning offspring’s immune system.
The preweaning colon has an influx of naive T cells, which can differentiate into long-lived dietary antigen–specific pTregs.
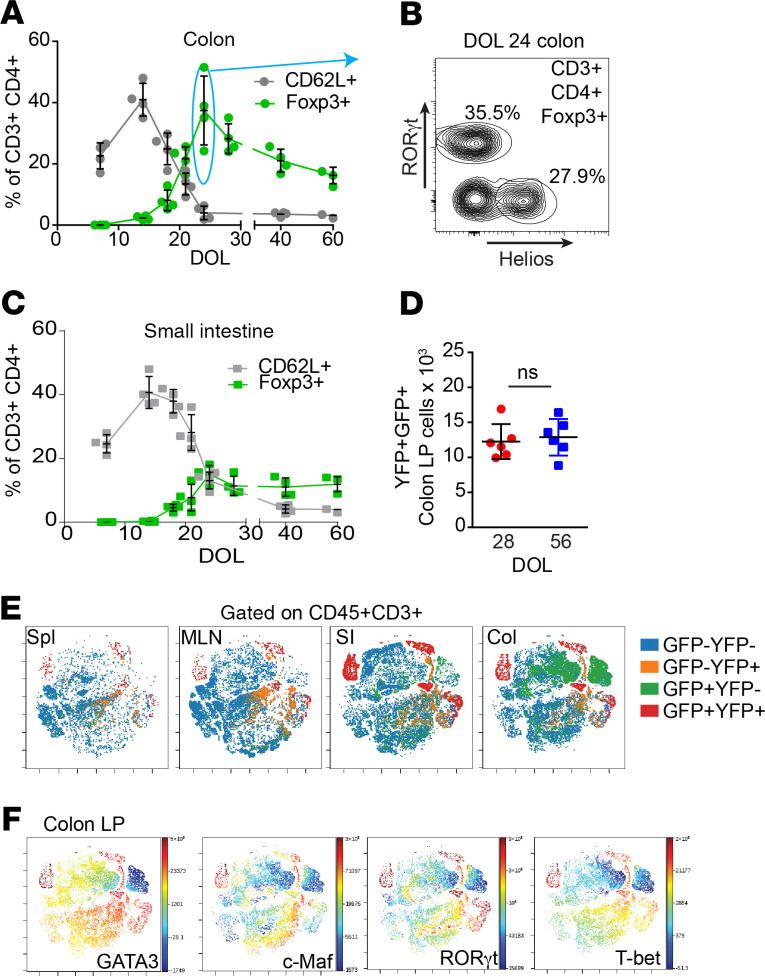
Naive T cells differentiate into peripherally derived Tregs (pTregs) that then mediate tolerance to environmental antigens. Loss or dysfunction of Tregs is associated with exaggerated immune responses, including Th2 responses characteristic of allergy (13–15). We evaluated naive T cell and Treg populations in the intestine throughout early life. Naive T cells (CD62L+CD44–CD69–) appeared in the colonic LP coinciding with the presence of colonic GAPs (Figure 2A and Supplemental Figure 2, compare with Supplemental Figure 1). The colonic LP naive T cell population dramatically decreased in frequency at weaning and remained low through adulthood. Approximately 1 week after the naive T cells peaked, a substantial population of Tregs appeared in the colonic LP (Figure 2A). The majority of these Tregs did not express Helios, the currently best available marker for thymus-derived Tregs (tTregs; Figure 2B), and approximately half of the Helios– Tregs expressed RORγt (Figure 2B). This is consistent with prior studies that have identified that RORγt+ pTregs appear in the gut beginning after 2 weeks of life (15, 46). In contrast to these studies, which observed that a population of colonic RORγt+ pTregs continues to expand through at least the first year of life (15), we observed that the population of RORγt+ pTregs did not expand after weaning but persisted in the colon of adults (Figure 2A and Supplemental Figure 2B). This difference in the ability to generate RORγt+ pTregs after weaning may in part be attributable to differences in the gut microbiota, as the development of RORγt+ pTregs after weaning can be driven by Helicobacter species (47, 48), which are largely absent from our colony (not shown). We found very few RORγt+Foxp3– cells in the colon (Supplemental Figure 2C), suggesting that RORγt expression is not a generalizable property of effector T cells generated during this time in life. A similar pattern of naive T cells was also observed in the SI LP; however, the peak in the Treg population in the SI LP was less pronounced (Figure 2C). The kinetics of naive T cell appearance, regression, and appearance of Tregs in the colonic LP appeared consistent with the kinetics of cognate antigen–driven conversion of naive T cells to pTregs (49), suggesting that these pTregs may have been induced in the colon in response to luminal substances and potentially dietary antigens.
Figure 2. A Long-lived diverse Treg population develops in the colon during a pre- and periweaning period.
(A) Naive (CD62L+) T cell and Treg (Foxp3+) populations in the colon LP in early life assessed by flow cytometry. (B) Helios and RORγt+ expression by DOL24 Foxp3+CD4+CD3+ colonic LP cells. (C) Naive (CD62L+) T cell and Treg (Foxp3+) populations in the SI LP in early life assessed by flow cytometry. (D) Absolute number of colonic Foxp3+ (GFP+) cells that express YFP on DOL28 or DOL56 following inducible expression of YFP on DOL24 in Foxp3GFPERT2CreROSAlslYFP mice. (E) viSNE plots of T cells from spleen (spl), mesenteric lymph node (MLN), SI, or Colon LP of DOL56 Foxp3GFPERT2CreROSAlslYFP mice following inducible expression in DOL24 analyzed by GFP and YFP. (F) viSNE plots demonstrating transcription factor expression by colon LP T cells isolated and clustered as in E. Data are presented as the mean ± SEM. *P < 0.05; each dot represents 1 mouse analyzed. B is representative of 1 of 4 mice analyzed; n = 4 or more mice in panels E and F; and offspring from 3 litters were used in A–F. Significance calculated using a 2-tailed Student’s t test in D.
Because the colon, as opposed to the SI, has been shown to be a site for the development of pTregs before and near weaning, and because the pTregs developing during this period have unique properties to shape subsequent immune responses, subsequent studies focused on colonic pTregs. We used Foxp3GFPERT2CreROSAlslYFP mice to label Tregs to identify the phenotypes and fates of Tregs that were generated before and around the time of weaning. The Tregs labeled to express YFP on DOL24 demonstrated stable Foxp3 expression and their numbers in the colon LP did not significantly change for at least a month (Figure 2D). They were also present to some extent in extraintestinal tissues (Figure 2E). Tregs can express a variety of transcription factors, which can impart functional capacities to effectively suppress their effector T cell counterparts (50–52). We observed that the colon pTregs developing before DOL24, GFP+YFP+ populations, expressed RORγt, c-Maf, T-bet, or GATA3 (Figure 2, E and F). These transcription factors were expressed to a lesser extent in the colonic Tregs developing after DOL24, GFP+YFP– populations (Figure 2, E and F). Tregs from the spleen, MLN, and SI also displayed expression of these transcription factors (Supplemental Figure 3). The pTregs developing after weaning in response to dietary antigens support tolerance; however, these pTregs appear to be more transient when compared with those generated in the preweaning colon (29). These data suggest that, during this preweaning interval, the colon may be a site for the development of a population of longer-lived pTregs.
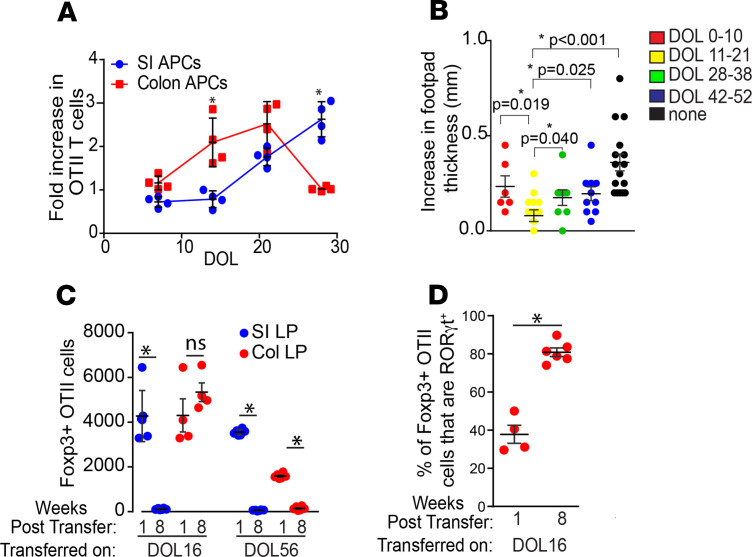
Prior work has shown that dietary antigen–specific pTregs are largely induced in the SI (29). However, these studies evaluated mice after weaning — a time when we observed that GAPs are present in the SI and largely absent from the colon (Supplemental Figure 1), which could limit dietary antigen delivery to the colonic immune system. Moreover, due to immature digestive mechanisms (53), dietary proteins could reach the colon in quantities that are sufficient to induce antigen-specific T cell responses in the preweaning gut but not in the more fully developed gut after weaning. Ovalbumin (Ova) is a food allergen and commonly used model antigen, which is effectively delivered to nursing mice via breast milk (54). We placed mice on dietary Ova and isolated APCs from the SI and colon weekly for the first 4 weeks of life. We observed that the expansion of Ova-specific OTII T cells cocultured with SI and colonic APCs isolated from mice receiving dietary Ova through early life mirrored the regional and temporal pattern of GAP formation, with colonic APCs being significantly more effective at inducing T cell expansion on DOL14 and with SI APCs being significantly more effective at inducing T cell expansion after weaning on DOL28 (Figure 3A compare with Supplemental Figure 1). This pattern could not be attributed to intrinsic differences in the antigen presentation capacity of the APCs because APCs from the SI and colon isolated at these times in early life induced similar OTII T cell expansion when exogenous Ova was added to the cultures (Supplemental Figure 4). To evaluate if introduction of a dietary antigen during this preweaning interval resulted in tolerance, we gave mice dietary Ova during this interval, DOL11–21, as well as similar 10 day intervals before and after this preweaning period, and evaluated tolerance using a delayed type hypersensitivity (DTH) assay. We found that introduction of dietary Ova during the 10-day interval when dietary antigen was largely delivered to the colonic immune system resulted in tolerance (Figure 3B). While we did observe significantly less footpad swelling in mice receiving dietary Ova from DOL11–21 when compared with all other groups (Figure 3B), it is not possible to control for the amount of dietary antigen delivered to the immune system or other events that may differ during these different intervals that may also contribute to this effect. However, our observations indicate that introduction of dietary antigen during the time when luminal antigen is delivered to the colonic immune system resulted in tolerance that was at least comparable to, if not slightly better than, tolerance to dietary antigen introduced at other times in life (Figure 3B).
Figure 3. Dietary antigen is delivered to the colonic immune system during a preweaning period, results in the induction of tolerance, and generates a population of long-lived dietary antigen–specific pTregs.
(A) Luminal antigen presentation capacity of SI and colon LP-APCs isolated from mice receiving dietary Ova in early life as assessed by the increase in Ova-specific OTII T cells after 72 hours of coculture. (B) Dietary Ova was given to mice for 10-day intervals throughout life, and tolerance was evaluated following Ova immunization and challenge, as assessed by footpad swelling. (C) Absolute number of Foxp3+ Ova-specific OTII cells in the SI or colon LP 1 or 8 weeks following transfer into DOL16 or DOL56 mice that were given Ova in drinking water for 7 days. (D) Percentage of colonic OTII T cells that are Foxp3+ and RORγt+ 8 weeks following transfer into DOL16 mice and 1 week of Ova in drinking water. Data are presented as the mean ± SEM, *P < 0.05; each point represents an individual mouse. Offspring were from 2 litters per condition and/or time point. Significance was calculated using 2-tailed Student’s t test in A, C, D. One-way ANOVA with a Dunnett’s post hoc test was used in B.
The above observations suggest that, during a pre- and periweaning period, dietary antigen–specific pTregs may be induced in the colon and that these dietary antigen–specific pTregs may be relatively long lived. Therefore, we isolated naive Ova specific T cells from OTII Rag–/– mice, which ensures sole expression of one TCR specific for Ova, and adoptively transferred them into preweaning mice or adult mice receiving dietary Ova for 1 week, and we evaluated the phenotypes and fates of the transferred cells 1 week and 7 weeks later. OTII T cells adoptively transferred to preweaning mice and adult mice receiving dietary Ova in early life became pTregs that localized to the SI and the colon LP at 1 week after transfer (Figure 3C). After 7 weeks off dietary Ova, the Ova-specific pTregs generated after weaning significantly regressed, while, in comparison, those generated before weaning persisted in the colon in the absence of cognate antigen (Figure 3C). The capacity to generate or maintain these long-lived dietary antigen–specific pTregs may be a unique feature of the preweaning colon, as the Ova-specific pTregs generated before weaning significantly regressed and did not persist in the SI to an appreciable extent in mice receiving OTII T cells on DOL16 (Figure 3C). The OTII T cells persisting in the colon became even more enriched in the expression of the transcription factor RORγt after 7 weeks off of dietary Ova (Figure 3D), consistent with the phenotype of the endogenous pTreg population generated in the preweaning colon.
Desynchronizing dams and pups disrupts colonic pTreg development:
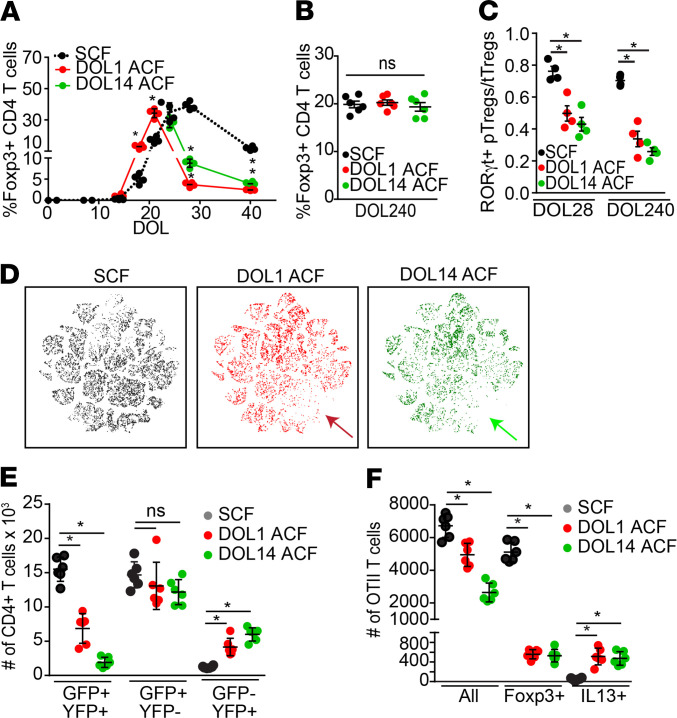
An inflammatory response to the gut microbiota, termed the weaning reaction, inversely correlates with pathological imprinting of the immune system, which can be phenocopied by loss of RORγt+ Tregs (27). Elevations of TNF-α and IFN-γ in the terminal ileum at DOL21 are reliable markers of the weaning reaction (27). Desynchronization of dams and pups altered the weaning reaction (27); however, the effects of desynchronization on the development of immune pathologies or on the RORγt+ pTreg population have not been explored. We did not observe the weaning reaction, evidenced by a significant increase in TNF-α or IFN-γ expression in the terminal ileum at the time of weaning, in mice in our colony, nor in mice with a more or less diverse gut microbial communities (Supplemental Figure 5A), suggesting that an overt inflammatory response to the gut microbiota at the time of weaning may not be a feature seen in all vivaria. We did observe that mice with a richer gut microbial community had a significantly larger colonic Treg population on DOL21 (Supplemental Figure 5B), consistent with microbes facilitating the development of colonic Tregs. We evaluated if desynchronization affected the colonic pTreg population. In SCF mice, the size and kinetics of the colonic LP Treg populations resembled the endogenous Treg population of unmanipulated animals (Figure 4A compare with Figure 2A). Mice in which colonic GAPs were inhibited after DOL14 (i.e., DOL14 ACF pups exposed to dams 1 day pp) undergo a significant diminution in their colonic Treg population following ACF (Figure 4A). While DOL1 ACF mice fostered to dams that had delivered 2 weeks earlier had a somewhat earlier peak and significantly increased colonic CD4+Foxp3+ T cell population before weaning, this population was not sustained and was significantly diminished after weaning (Figure 4A). Why the Foxp3+ population was not maintained in DOL1ACF mice is unclear but could reflect transient and promiscuous expression of Foxp3 by activated or effector T cells, as has been previously described (55–57). By DOL240, the total colonic LP Treg populations in the ACF mice were not significantly different when compared with the SCF mice (Figure 4B); however, the colonic RORγt+ pTreg population remained significantly diminished for at least 8 months, the last time point evaluated (Figure 4C). These deficits in DOL1 and DOL14 ACF mice affected similar Treg populations expressing RORγt, c-Maf, GATA3, or T-bet (Figure 4D and Supplemental Figure 6). Using the Foxp3GFPERT2CreROSAlslYFP mice, we performed fate mapping studies of cross-fostered mice whose Tregs were labeled on DOL24 and analyzed on DOL56. ACF mice had significantly reduced Tregs generated before weaning (GFP+YFP+ cells) and had significantly more T cells that had lost Foxp3 expression (GFP–YFP+ cells), with DOL14 ACF mice being more severely affected (Figure 4E). The population of colonic cells expressing Foxp3GFP after DOL24 (GFP+YFP– cells) was not significantly affected by ACF (Figure 4E). We treated mice with inhibitors of EGFR activation (EGFRi) before DOL10, to mimic effects of exposure to lower levels of EGF seen in DOL1 ACF mice, or we gavaged mice with EGF from DOL10–21, to mimic effects of higher exposure to EGF seen in DOL14 ACF mice. Similar to the DOL1 ACF mice, which are exposed to lower levels of EGF, mice treated with EGFRi before DOL10 had an earlier peak in the colonic CD4+Foxp3+ population, which was significantly larger on DOL21; however, this was not maintained and became significantly decreased after weaning (Supplemental Figure 7A). Mice treated with EGF from DOL10–21 to inhibit GAPs and antigen delivery to the colonic immune system had a significantly smaller colonic CD4+Foxp3+ population on following EGF treatment (Supplemental Figure 7A). This is consistent with the hypothesis that deficits in the Treg population resulting from ACF are caused, at least in part, from altered EGF exposures.
Figure 4. Desynchronizing pups and dams impairs the development of a long-lived population of colonic Tregs that can be specific for dietary antigen.
(A) Population of colonic LP Tregs in cross-fostered mice by DOL. (B) Total Treg population in cross-fostered mice on DOL240. (C) Ratio of RORγt+ pTregs (Helios–) to tTregs (Helios+) in the colonic LP of cross-fostered mice at DOL28 or DOL240. (D) viSNE plots of colonic LP T cells isolated from cross-fostered mice on DOL80 stained for transcription factors as in Supplemental Figure 6; arrows denote missing population following ACF. (E) Number of colon LP CD4+ T cells expressing YFP and or GFP in DOL56 cross-fostered Foxp3GFPERT2CreROSAlslYFP mice following inducible expression on DOL24. (F) Absolute number of Ova-specific OTII T cells in the colon LP of cross-fostered mice receiving OTII T cells on DOL16, given Ova in drinking water, and evaluated 1 week later. Data are presented as the mean ± SEM, *P < 0.05; each dot represents analysis of 1 mouse. n = at least 4 mice per group; offspring from 3 litters per condition and/or time point were used in A–F. D is representative of 3 independent pooling analysis with 2 mice per group. Significance calculated using 2-way ANOVA with a Dunnett’s post hoc test in A and 1-way ANOVA with a Dunnett’s post hoc test in B, C, E, and F.
To evaluate if pTregs specific for dietary antigen were affected by ACF, we transferred Ova-specific naive CD4+ OTII T cells into DOL16 cross-fostered mice receiving Ova in drinking water and evaluated the phenotype of the transferred cells. OTII expansion was significantly reduced in both groups of ACF mice, with DOL14 ACF being more affected (Figure 4F). ACF affected the phenotype of the OTII cells, as the number of Foxp3-expressing OTII T cells was significantly reduced in both groups of ACF mice (Figure 4F). This effect to significantly reduce the population of Foxp3+ Ova-specific T cells could be recapitulated by inhibiting EGFR activation before DOL10, to mimic DOL1 ACF, or by EGF gavage from DOL10–21, to mimic DOL14 ACF (Supplemental Figure 7B), consistent with the interpretation that alterations in EGF exposure in the preweaning gut can affect the generation of dietary antigen–specific Tregs. A small but significant proportion of OTII T cells in both ACF mice groups expressed IL-13 when compared with the SCF mice (Figure 4F), suggesting that ACF may induce Th2 responses to dietary antigens encountered before weaning.
Desynchronization results in skewing of the immune system and impaired tolerance to newly encountered dietary antigens.
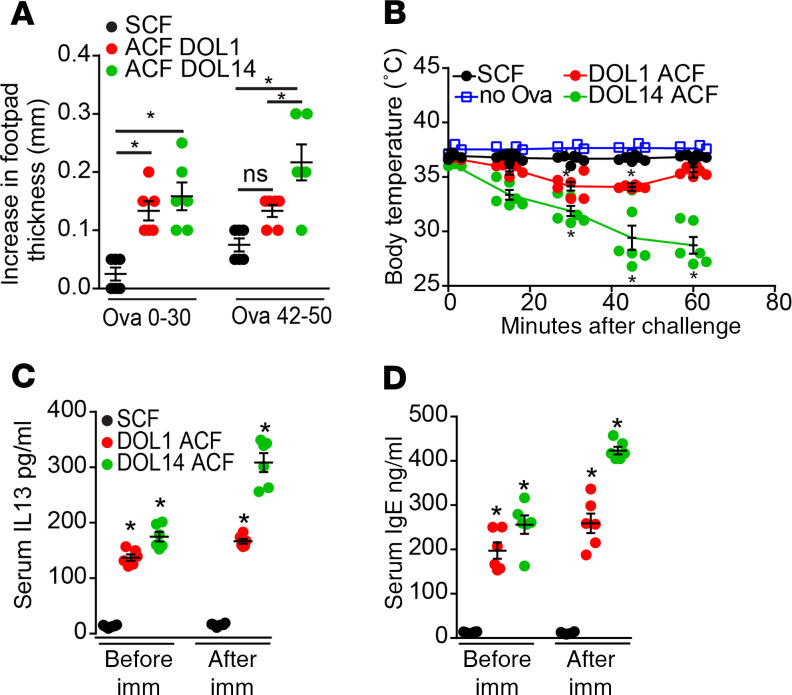
The above data suggest that mothers and their offspring are synchronized to facilitate the development of a long-lived pTreg population, which can be specific for dietary antigens and could support oral tolerance. To confirm this, we synchronously or asynchronously cross-fostered mice and used a DTH assay to evaluate tolerance to dietary antigen initially introduced during and around the time of cross-fostering, as well as dietary antigens initially encountered later in life. DOL1 ACF mice and DOL14 ACF mice had reduced tolerance to dietary antigens introduced around the time of cross-fostering, as evidenced by significantly increased footpad swelling (Figure 5A). The impaired oral tolerance to dietary antigen introduced in early life is consistent with our observations that the ability to generate dietary Ova-specific Tregs following ACF is reduced (Figure 4F). Surprisingly, DOL1 ACF mice showed a trend toward a deficit — and DOL14 ACF mice displayed a significant deficit — in tolerance to dietary Ova that was introduced in adulthood (Figure 5A). Moreover, following immunization and footpad challenge with sterile Ova, DOL1 and DOL14 ACF mice became significantly hypothermic (Figure 5B) and lethargic (not shown), recapitulating features of a hypersensitivity reaction. Indeed, DOL1 ACF mice and DOL14 ACF mice had significant elevations of serum Th2 mediators and global IgE at baseline and after immunization, as well as significantly elevated antigen-specific IgE following immunization but did not have significant elevations in prototypic Th1 or Th17 cytokines when compared with SCF mice (Figure 5, C and D, and Supplemental Figure 8). In combination with the above findings, this suggests that the pTregs generated in the pre- and periweaning period act to promote tolerance to newly encountered antigens and to restrain Th2 responses.
Figure 5. Desynchronization of pups and dams results in impaired oral tolerance and exaggerated Th2 responses.
(A–D) Mice that were synchronously cross-fostered (SCF) or asynchronously cross-fostered (ACF) on DOL1 or DOL14 (ACF DOL1, ACF DOL14) were given Ova in drinking water on DOL0–30 or on DOL42–50 and immunized against Ova as adults. (A) Quantitative increase in footpad thickness following challenge with Ova in the footpad. (B) Change in body temperature following footpad challenge with Ova in mice receiving dietary Ova from DOL0–30. (C and D) Serum (C) IL-13 and (D) IgE before immunization or after immunization and footpad challenge. Data are presented as the mean ± SEM, *P < 0.05. A–D is representative of 3 independent experiments with 4 or more mice in each group; offspring from 3 litters per condition were used in A–D. Significance calculated using 1-way ANOVA with a Dunnett’s post hoc test in A, C, and D, and 2-way ANOVA with a Dunnett’s post hoc test in B.
pTregs from the periweaning colon rescue immune skewing and impaired tolerance to dietary antigen.
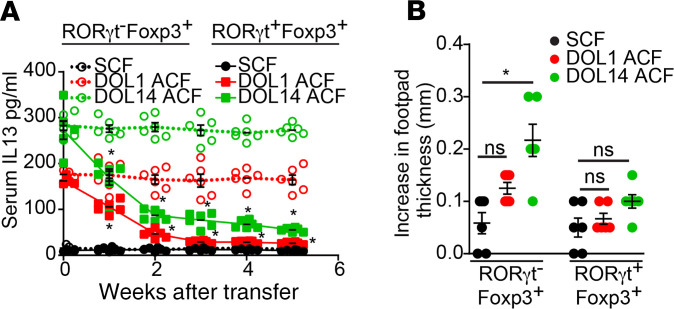
ACF could have effects contributing to impaired tolerance and immune skewing beyond the loss of the colonic pTreg population. To evaluate if these colonic pTregs were sufficient to rescue impaired tolerance induction and the Th2 skewing resulting from ACF, we adoptively transferred 5 × 104 Foxp3+RORγt+ Tregs isolated from the colonic LP of DOL21 mice or 5 × 104 Foxp3+RORγt– Tregs isolated from the spleen, MLN, and colonic LP of DOL21 mice into DOL56 mice that had been cross-fostered and evaluated serum levels of IL-13 and the ability to induce tolerance to dietary antigens. The elevated serum concentrations of IL-13 seen in ACF mice decreased significantly 2 weeks after transfer of the RORγt+ Tregs isolated from the periweaning colon and remained low 5 weeks after transfer, the latest time point evaluated (Figure 6A). However, transfer of the RORγt– Tregs did not significantly affect the elevations in IL-13 in ACF mice (Figure 6A). This indicates that the periweaning colonic pTregs can have effects to help maintain immune homeostasis. Moreover, transfer of RORγt+ colonic Tregs, but not RORγ– Tregs, isolated from DOL21 mice rescued the impaired tolerance induction to dietary Ova introduced in adulthood in DOL14 ACF mice (Figure 6B), thus demonstrating that restoring this Treg population was sufficient to reverse Th2 skewing and impaired tolerance induction following ACF.
Figure 6. Tregs from the periweaning colon can rescue oral tolerance and immune skewing in desynchronized pups.
(A and B) A total of 5 × 104 Foxp3+RORγt+ Tregs isolated from DOL21 colon LP or 5 × 104 Foxp3+RORγt– Tregs isolated from DOL21 spleen, MLN, and colon LP were transferred into DOL56 cross-fostered mice as described in Figure 5. (A) Serum IL-13 concentrations measured weekly after transfer. (B) On DOL98, mice were given Ova in drinking water for 1 week, immunized with Ova, and challenged with Ova in the footpad. The increase in footpad thickness was measured 24 hours later. Data are presented as the mean ± SEM, *P < 0.05. A and B are representative of 2 independent experiments with 5 or more mice in each group; offspring from 3 litters per condition were used in A and B. Significance was calculated using 2-way ANOVA with Dunnett’s post hoc test in A and 1-way ANOVA with a Dunnett’s post hoc test in B.
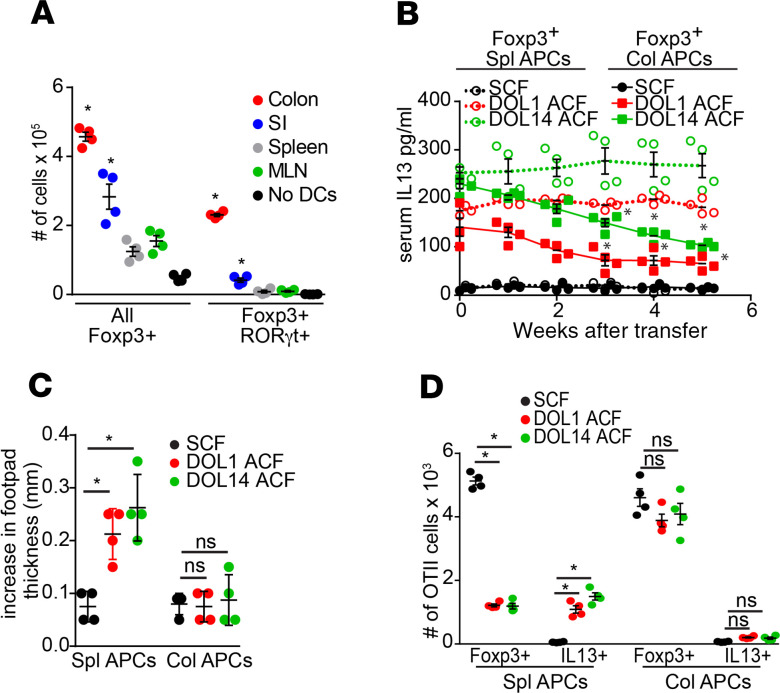
We observed that naive OTII splenic T cells from adult mice adoptively transferred into DOL16 mice could become long-lived RORγt+ colonic Tregs (Figure 3C), indicating that naive T cells from adult mice, if placed in the correct environment, could recapitulate events seen in preweaning mice and suggesting that, during this preweaning interval, the colon may have unique properties supporting the generation of this pTreg population. Prior studies have shown that RORγt+Foxp3+ Tregs can be generated using in vitro culture of naive T cells stimulated with anti-CD3, anti-CD28, and cytokine cocktails; however, these induced Tregs (iTregs) did not fully recapitulate the functions of their in vivo pTreg counterparts (58). We explored if an iTreg with the capacity to rescue the phenotype seen in ACF could be generated in vitro from preweaning colonic LP-APCs and naive T cells. Indeed, colonic LP-APCs from DOL21 mice had a significantly enhanced capacity to generate iTregs in vitro in general and specifically RORγt+ iTregs when compared with SI-LP, spleen, or MLN APCs isolated from the same mice (Figure 7A). As with the endogenous population of pTregs from the DOL21 colon, adoptive transfer of these iTregs significantly reduced the Th2 skewing, rescued the impaired oral tolerance induction to dietary antigen, and facilitated the induction of Tregs to newly encountered dietary antigens in ACF mice (Figure 7, B–D).
Figure 7. In vitro–generated Tregs using periweaning colonic APCs rescues Th2 skewing and oral tolerance in desynchronized pups.
(A) Naive splenic T cells (RFP–GFP–) isolated from Foxp3RFPRORγtGFP mice were cultured for 7 days with MHCII+CD11c+ cells isolated from colon, SI, spleen, or MLN of DOL21 mice and analyzed by flow cytometry for Foxp3 and RORγt expression. (B–D) A total of 5 × 104 Foxp3+ cells generated by culture with DOL21 colon LP-APC or by culture with DOL21 splenic APCs was transferred into DOL56 cross-fostered mice. (B) Serum IL-13 concentrations measured weekly after transfer. (C) Increase in footpad thickness following dietary Ova, Ova immunization, and footpad challenge in mice treated as in B. (D) Absolute number of Foxp3+ or IL13+ Ova-specific OTII T cells 7 days following transfer into mice treated as in B and given dietary Ova. Data are presented as the mean ± SEM, *P < 0.05. A is representative of 1 of 3 independent experiments with n = 3 replicate culture wells, B–D is representative of 2 independent experiments with 4 or more mice in each group, and offspring from 3 litters per condition were used in A–D. Significance was calculated using 1-way ANOVA with a Dunnett’s post hoc test in A, C, and D and 2-way ANOVA with a Dunnett’s post hoc test in B.
Discussion
Our observations give insight into prior studies identifying the benefits of breastfeeding, the complementary introduction of food allergens, and early exposure to gut microbes for reducing allergic outcomes (22, 26, 30, 32, 36, 59, 60). These time-restricted benefits are, in part, linked to the development of a population of pTregs in early life. Studies of germ-free and antigen-free mice indicate that pTregs induced to foods after weaning are continuously generated in the SI and recede when food antigens are removed (29). In contrast, our findings suggest that the pTregs generated when breast milk allows antigen delivery to the colonic immune system during a defined interval can be specific for commensal bacterial antigens (40) or dietary antigens (presented here); are longer-lived; and have capacities to promote tolerance to dietary antigens encountered during this interval, to promote tolerance to dietary antigens encountered in the future, and to control Th2 responses. The benefit of antibiotic avoidance in the first year of life directly aligns with our observations, as multiple studies have identified a role for gut microbes and microbial products in driving the development of gut Tregs (15, 18, 19, 46). Hence, the above practices may have at least 2 benefits. First, introducing a food allergen during this preweaning interval generates long-lived food allergen–specific Tregs, thereby conferring a sustained tolerance to that allergen. The longevity of these pTregs, compared with those generated in response to a dietary antigen after weaning (observations presented here and in ref. 29) may, in part, explain why continual dietary antigen exposure is not required to reduce food allergy in at-risk children when food allergens are introduced early (36). Second, breastfeeding that is synchronized between mothers and offspring and antibiotic avoidance promotes the development of a long lived pTreg population with broad specificities for dietary and gut commensal antigens and may enforce immune homeostasis, promote tolerance to new dietary antigens, and tip the balance away from allergy in at-risk children.
Prior observations and observations presented here suggest that the colonic RORγt+ pTregs generated before and near weaning have properties that differ from those generated after weaning (27). While we observed that the RORγt+ colonic pTreg population was not restored in later life, others have found that colonic RORγt pTregs can continue to develop after weaning (15). The differences between in the ability to generate colonic RORγt+ pTregs after weaning between these studies and ours likely reflects differences in the gut microbial communities with an absence of bacterial taxa with the capacity to generate colonic RORγt+ Tregs after weaning in our mice. We do not believe the generation of colonic RORγt+ pTregs after weaning would reverse the immune skewing we observed in desynchronized mice. This is in agreement with studies demonstrating that deletion of all Tregs in the periweaning period resulted in immune pathology later in life, and prior observations from this group demonstrating that RORγ+ pTregs were continuously generated for at least the first year of life in mice in their colony (15, 27).
By extension the above observations might also suggest that bacterial taxa and/or the properties of the bacterial taxa driving the generation of the pre- and periweaning colonic pTregs differ from those driving the generation of postweaning colonic pTregs. Bacteria that adhere to the gut epithelium can be continually encountered by the immune system in manners independent of GAPs and can drive the development of colonic bacterial antigen–specific RORγt+ pTregs after weaning (47, 48, 61–63). Conversely, some commensal bacteria are encountered by the immune system via colonic GAPs before weaning and drive the development of commensal bacterial antigen–specific pTregs (40). During homeostasis, these commensal bacteria are largely unseen by the immune system outside of this preweaning interval but can be reencountered during barrier breach or infection (40, 64). Collectively, these observations suggest colonic RORγt+ pTregs generated in response to adherent bacteria after weaning do not share all of the properties of colonic RORγt+ pTregs generated before and near weaning and are not able to compensate for their absence.
An inflammatory response to the gut microbiota around the time of weaning, or the weaning reaction, has been found to correlate with the development RORγt+ pTregs and the prevention of pathological immune imprinting (27). The weaning reaction and the development of RORγt+ pTregs are both driven by the gut microbiota (15, 27, 45); accordingly, defining the role of the weaning reaction, as opposed to the role of gut microbes independent of this inflammatory response, on the development of colonic RORγt+ pTregs and the prevention of pathological immune imprinting is complex. Monocolonization of germ-free mice with segmented filamentous bacteria (SFB) induced the weaning reaction but did not protect from pathological imprinting (27), indicating that this overt inflammatory response to gut microbiota is not always sufficient to protect from pathological imprinting. That SFB produces an inflammatory response in the ileum around weaning in not completely unexpected, as SFB colonizes the ileum and IgA from maternal milk suppresses SFB colonization until weaning (65, 66). Unmanipulated mice in our colony did not display pathological immune imprinting and lacked an overt inflammatory response to the microbiota at the time of weaning; nonetheless, an immune response to the microbiota around weaning has been found to be critical for the development of a balanced immune system (27, 67).
We observed that desynchronization resulted in loss of colonic RORγt+ pTregs developing in the pre- and periweaning interval, spontaneous Th2 skewing, and impaired tolerance to new dietary antigens. These deficits could be corrected by adoptive transfer of colonic RORγt+ pTregs from periweaning mice into adult mice that had been desynchronized as pups. This demonstrates that these colonic RORγt+ pTregs were sufficient to correct these defects. While mice desynchronized as pups appeared normal in absence of challenge, these observations imply a role for these pTregs to maintain balanced immunity in later life, which is in line with our observations of fate-mapping studies of Tregs developing near weaning. While mouse models cannot fully recapitulate human disease, our observation that adoptive transfer of syngeneic Tregs generated in vitro can suppress an allergic phenotype in mice gives hope that this approach could translate to humans with refractory allergic disorders.
Findings presented here suggest that breastfeeding is protective of an allergic phenotype. While this is in line with many epidemiological studies, the benefit of breastfeeding to prevent allergic outcomes in humans remains unclear, in part, due to the heterogenous nature of observational studies and the inability to perform randomized trials of breastfeeding in children due to ethical concerns. Furthermore, our results suggest that the benefit of breastfeeding could be most apparent in offspring receiving synchronized breastmilk, which raises new complexities in how feeding practices are classified and how to interpret observational studies correlating feeding practices and subsequent outcomes. Premature infants in the neonatal intensive care unit (NICU) are often fed donor milk, which may not be synchronized with the infant, when their mother’s own milk is not available. These infants are at risk for multiple adverse outcomes; however, the current data on feeding practices of infants in the NICU are not granular enough to correlate desynchronization with these outcomes. Therefore, while this is a sizeable real-world example of desynchronization in children at risk for multiple adverse outcomes, it is currently not possible to give priority to the contribution of desynchronization versus the many other risk factors these children encounter, on these outcomes.
We acknowledge that our study was largely performed on inbred mice in a controlled vivarium, whether these observations directly translate to humans, who have a greater genetic diversity and richer gut microbiota, is not readily apparent. However, studies demonstrating the benefit of complementary breastfeeding, early food allergen introduction, and antibiotic avoidance in the first year of life on allergic outcomes suggest that similar physiology is occurring in mice and humans. An additional limitation of our work is that we focused on the effect of desynchronization on the development of colonic pTregs, and the effects of desynchronization may extend well beyond this cellular population and effects of alterations in EGF exposure. Indeed, recent studies have found that breast milk IgA levels inversely correlate with colonic RORγt+ Tregs in the offspring and that this is a phenotype transmissible across generations (68). Future work will attempt to correlate allergic outcomes with desynchronization in humans and examine the effects of desynchronization beyond EGF and this cellular population.
Our observations align with recent studies supporting that early introduction of food allergens can reduce food allergy in at-risk children. However, avoiding practices that disrupt the development of this population of pTregs, such as desynchronization and altering in the gut microbiota with antibiotics during this interval, will have even greater consequences. Hence, the short-term benefit of antibiotics in preweaning children should be weighed against the longer-term risk of allergies. Additionally, our data raise concerns about the use of feeds that are not synchronized with the developing infant. In concert, these practices may contribute to the increase in allergic diseases seen in populations worldwide.
Methods
Supplemental Methods are available online with this article.
Mice.
All mice were maintained on the C57BL/6 background. C57BL/6 mice, B6SJL mice, OTII T cell receptor transgenic mice (69), Math1fl/fl mice (70), FoxP3GFPCreERT2 mice (56), RosalslYFP mice (71), Foxp3RFP mice (72), and RORγtGFP mice (73) were purchased from The Jackson Laboratory. OTII Rag–/– T cell receptor transgenic mice (69) were purchased from Taconic Farms. Transgenic mice bearing a tamoxifen-inducible Cre recombinase expressed under the control of the villin promoter (vil-Cre-ERT2) mice (74) were a gift from Sylvie Robine (Institut Curie, Paris, France). Germ-free mice were obtained from the Washington University Digestive Disease Research Core Center. Pregnant murine pathogen free and excluded flora C57BL/6 mice were purchased from Taconic Biosciences and housed separately. All mice were fed a routine chow diet. Mice other than those with defined flora were bred and maintained in house. Cohoused littermates were used for experimental controls where possible. All mice were weaned at DOL21. Adult mice were 8–16 weeks of age at the time of analysis unless stated otherwise.
Oral tolerance and DTH responses.
Mice and/or nursing mothers when treated before weaning, were given Ova 20 g/L in drinking water or drinking water alone for the time periods indicated in the figures. Mice receiving Ova for 10-day intervals before weaning (Figure 3B) were immunized s.c. with 100 μg Ova in incomplete Freund’s Adjuvant (MilliporeSigma) at 6 and 8 weeks of age. Mice receiving Ova for 10-day intervals after weaning (Figure 3B) were immunized on a similar schedule starting 21 days after the last exposure to dietary Ova. Fifteen weeks following the initial exposure to dietary Ova, mice were challenged with 20 μg Ova in the footpad, and 24 hours later, the DTH response was read as the increase in footpad thickness as compared between before and after challenge, measured with micrometer calipers. For all other DTH experiments in cross-fostered mice, mice were immunized 2 weeks and 4 weeks following the last exposure to dietary Ova and challenged with 20 μg Ova in the footpad 2 weeks later. In some experiments, body temperature was monitored for 1 hour following challenge.
Analysis of luminal antigen capture by LP-APCs.
To evaluate luminal antigen presentation capacity, mice were given Ova 20 g/L (MilliporeSigma) for 3 days before isolation of LP-APCs (dietary Ova, Figure 3A). LP-APC populations and Ova-specific CD4+ OTII T cells were isolated with flow cytometric cell sorting and cultured at a ratio of 1:10 APCs to T cells as previously described (75, 76). As a positive control, 10 μg Ova was added to cultures of APC populations isolated from mice not receiving dietary Ova. After 3 days, cultures were evaluated for the number of T cells by flow cytometry and cell counting.
Adoptive T cell transfer of Ova-specific T cells.
Nursing (CD45.1) B6.SJL dams with DOL14 pups or 8-week-old B6.SJL females were given Ova 20 g/L in drinking water for 7 days, and 2 days after the start of dietary Ova, mice were injected i.p. with 5 × 105 flow cytometrically sorted naive Ova-specific T cells (CD45.2+CD3+CD4+CD62L+Vα2+) isolated from the spleen of adult OTII Rag–/– mice. Following transfer, organs were harvested at the indicated time points for cell isolation and analyzed by flow cytometry to detect and evaluate the phenotype of the CD45.2+ OTII T cells. In some experiments, 5 × 105 FACS isolated CD45.1+CD3+CD4+CD62L+Vα2+ OTII T cells from the spleen of an adult OTII transgenic mouse on the congenic CD45.1 background was injected i.p. on DOL16 into SCF mice, DOL1 ACF mice, or DOL14 ACF mice receiving Ova in drinking water between DOL14 and DOL21. Seven days following transfer (DOL23), organs were harvested for cell isolation and analyzed by flow cytometry to detect the CD45.1+ OTII T cells. In rescue experiments evaluating the phenotype of dietary antigen–specific T cells, SCF mice, DOL1 ACF mice, or DOL14 ACF received Ova in drinking water from DOL70–77 and were injected i.p. on DOL72 with 5 × 105 FACS isolated CD45.1+CD3+CD4+CD62L+Vα2+ OTII T cells from the spleen of an adult OTII transgenic mouse. Mice were evaluated 7 days after transfer for the phenotype of the transferred CD45.1+ OTII T cells.
Rescue of ACF mice.
A total of 5 × 104 CD45+CD3+CD4+Foxp3+RORγt+ cells were FACS sorted from the colon or 5 × 104 or CD45+CD3+CD4+Foxp3+RORγt– cells were FACS sorted from the spleen, MLN, and colon of DOL21 Foxp3RFPRORγtGFP mice and injected i.p. into SCF, DOL1 ACF, and DOL14 ACF mice on DOL56. Alternatively, CD45+CD3+CD4+CD62L+ FACS-sorted splenocytes isolated from Foxp3RFPRORγtGFP adult mice (6–8 weeks of age) were cultured in vitro in a 1:1 ratio with FACS-sorted CD11c+MHCII+APCs isolated from the spleen, MLN, SI, or colon of DOL21 mice using conditions as previously described (41, 76). Seven days later, cells were analyzed for Foxp3RFP and RORγtGFP expression by flow cytometry, and 5 × 104 cells were injected i.p. into SCF, DOL1 ACF, and DOL14 ACF mice on DOL56. Mice were bled weekly to monitor serum IL-13. In some experiments, mice were then given Ova in the drinking water for 7 days, immunized with Ova, and challenged with Ova via footpad injection to analyze the ability to initiate oral tolerance. In some experiments, the phenotype of dietary antigen–specific T cells was evaluated with adoptive transfer of OTII T cells as above.
Enumeration of GAPs.
GAPs were identified and enumerated using in vivo 2 photon imaging and fluorescent microscopy on fixed tissue sections as previously described (41, 76).
Extraction of milk from lactating dams.
Milk was extracted from lactating dams every third day, beginning on day 5 pp as previously described (77).
Stool specimens.
Preexisting stool specimens with extensive linked metadata on feeding practices were used for this study. These stool specimens were collected from a cohort of twins followed at St. Louis Children’s Hospital. Data were collected regarding pregnancy, labor, delivery, medications, and feeding by reviewing medical records and interviewing parents. Stool specimens used for this study were obtained from healthy children delivered at > 35 weeks gestation who were not on continuous medication through the study. Selection of stool specimens for this study was based upon the absence of chronic illnesses or multiple courses of antibiotics and the presence of feeding history to assign to a group. Exclusive breastfeeding was defined as breastfeeding without formula feeding. Exclusive formula feeding was defined as formula feeding without breastfeeding. Complementary consumption of solid foods was not a criterion for exclusion for assignment to either group. Specimens from children receiving donor milk were not used in this study.
Measurement of cytokines, EGF, and immunoglobulins.
ELISAs specific for murine EGF (R&D systems), human EGF (R&D systems), IL-13 (eBioscience), Eotaxin (R&D systems), IgE (eBioscience), IFN-γ (eBioscience), TNF-α (eBioscience), IL-17 (eBioscience), and IL-10 (eBioscience) were used per the manufacturers recommendations. An ELISA consisting of Ova (MilliporeSigma, A5503) for capture, and monoclonal anti-IgE antibody (Southern Biotech, 1110-04) conjugated to alkaline phosphatase for detection was used to measure Ova-specific IgE. An ELISA consisting of monoclonal anti-IgG (Southern Biotech, 1030-01) and monoclonal anti-IgG conjugated to alkaline phosphatase (Southern Biotech, 1030-04) for detection was used to measure total IgG.
Statistics.
Data were assumed to be normally distributed. Analysis was performed using a 2-tailed Student’s t test or 1-way ANOVA with a Dunnett’s post hoc test with GraphPad Prism (GraphPad Software Inc.) and a cutoff of P < 0.05 for significance.
Study approval.
Animal procedures and protocols were performed in accordance with the IACUC at Washington University School of Medicine. Protocols used for recruitment of human participants, obtaining informed consent, collecting and deidentifying stool samples, and acquiring and deidentifying clinical metadata were approved by the Human Research Protection Office of Washington University School of Medicine. Consent was obtained from women with twin pregnancies to collect stool specimens from their twins (monthly to age 2 years and bimonthly thereafter).
Author contributions
KAK performed cell transfers, fate-mapping, in vitro cell studies, and data analysis. KAK, JKG, DHK, MG, and KGM performed animal models, 2P imaging experiments, and data analysis. KAK and PEC performed immunofluorescence, cell isolation, ELISAs, and data analysis. KAK and DHK performed viSNE analysis. KAK, BR, and VJ performed flow cytometry and gene expression. PIT, BBW, IMN, and AB managed the human studies and specimens. KAK, PIT, SPH, COE, CSH, and RDN designed the experiments, analyzed and interpreted the data, and wrote the manuscript. All authors reviewed and approved the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank Mark J. Miller for assistance with 2P imaging and analysis (MJM). The 2P imaging was performed at the In Vivo Imaging Core at Washington University School of Medicine. The Washington University Digestive Diseases Research Center Core, supported by NIH grant P30 DK052574, provided the germ-free mice. The High Speed Cell Sorter Core at the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, Missouri, USA — supported in part by NCI Cancer Center Support Grant P30 CA91842 — provided flow cytometric cell sorting services. The Speed Congenics Facility of the Rheumatic Diseases Core Center, supported by NIH grant P30AR048335 bred the Myd88–/– mice onto the C57BL/6 background. This work was supported by the following grants: DK097317 (RDN), AI131342 (RDN and PIT), AI140755, and AI136515 (RDN and CSH); AI112626 (SPH and RDN); DK071176 (COE); DK052574 and DK125606 (BR); DK109006 and AI095542 (KAK); K08DK101608, R03DK111473, and R01DK118568 (MG); March of Dimes Foundation grant 5-FY17-79 (MG); and the Children’s Discovery Institute grants MI-FR-2017-596 (MG) and MD-II-2018-725 (BBW).
Version 1. 08/06/2020
Electronic publication
Footnotes
KAK’s present address is: Department of Immunology, Mayo Clinic, Rochester, Minnesota, USA.
JKG’s present address is: Department of Neuroscience and Physiology, University of Gothenburg, Gothenburg, Sweden.
Conflict of interest: RDN, KAK, and KGM are inventors on U.S. Nonprovisional Application Serial No. 15/880,658 Compositions And Methods For Modulation Of Dietary And Microbial Exposure. PIT discloses a financial conflict of interest with MediBeacon Inc (member of their Scientific Advisory Board, consultant, and equity holder) and is a potential recipient of royalties from a patent to test human gut permeability noninvasively.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(15):e137943.https://doi.org/10.1172/jci.insight.137943.
Contributor Information
Devesha H. Kulkarni, Email: dkulkarn@wustl.edu.
Brigida Rusconi, Email: b.a.rusconi@wustl.edu.
Vini John, Email: vini.john@wustl.edu.
I. Malick Ndao, Email: ndao_m@wustl.edu.
Avraham Beigelman, Email: beigelmana@wustl.edu.
Misty Good, Email: mistygood@wustl.edu.
Charles O. Elson, Email: coelson@uab.edu.
Chyi-Song Hsieh, Email: chsieh@dom.wustl.edu.
References
- 1.Pawankar R. Allergic diseases and asthma: a global public health concern and a call to action. World Allergy Organ J. 2014;7(1):12. doi: 10.1186/1939-4551-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinberg EG. The WAO White Book on Allergy 2011-2012. Curr Allergy Clin Im. 2011;24(3) [Google Scholar]
- 3.Platts-Mills TA. The allergy epidemics: 1870-2010. J Allergy Clin Immunol. 2015;136(1):3–13. doi: 10.1016/j.jaci.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242(1):10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabieses B, Uphoff E, Pinart M, Antó JM, Wright J. A systematic review on the development of asthma and allergic diseases in relation to international immigration: the leading role of the environment confirmed. PLoS ONE. 2014;9(8):e105347. doi: 10.1371/journal.pone.0105347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ege MJ, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364(8):701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 7.Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clin Exp Immunol. 2010;160(1):1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbst T, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184(2):198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 10.Hill DA, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18(4):538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feehley T, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. 2019;25(3):448–453. doi: 10.1038/s41591-018-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefka AT, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci USA. 2014;111(36):13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458(7236):351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Josefowicz SZ, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482(7385):395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohnmacht C, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt+ T cells. Science. 2015;349(6251):989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 16.Stelmaszczyk-Emmel A, Zawadzka-Krajewska A, Szypowska A, Kulus M, Demkow U. Frequency and activation of CD4+CD25 FoxP3+ regulatory T cells in peripheral blood from children with atopic allergy. Int Arch Allergy Immunol. 2013;162(1):16–24. doi: 10.1159/000350769. [DOI] [PubMed] [Google Scholar]
- 17.Noval Rivas M, et al. Regulatory T cell reprogramming toward a Th2-cell-like lineage impairs oral tolerance and promotes food allergy. Immunity. 2015;42(3):512–523. doi: 10.1016/j.immuni.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 19.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metsälä J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Mother’s and offspring’s use of antibiotics and infant allergy to cow’s milk. Epidemiology. 2013;24(2):303–309. doi: 10.1097/EDE.0b013e31827f520f. [DOI] [PubMed] [Google Scholar]
- 21.Mitre E, Susi A, Kropp LE, Schwartz DJ, Gorman GH, Nylund CM. Association Between Use of Acid-Suppressive Medications and Antibiotics During Infancy and Allergic Diseases in Early Childhood. JAMA Pediatr. 2018;172(6):e180315. doi: 10.1001/jamapediatrics.2018.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love BL, Mann JR, Hardin JW, Lu ZK, Cox C, Amrol DJ. Antibiotic prescription and food allergy in young children. Allergy Asthma Clin Immunol. 2016;12:41. doi: 10.1186/s13223-016-0148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arrieta MC, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 24.Russell SL, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13(5):440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4(2):158–164. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336(6080):489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al Nabhani Z, et al. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity. 2019;50(5):1276–1288.e5. doi: 10.1016/j.immuni.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol. 2012;5(3):232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim KS, et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science. 2016;351(6275):858–863. doi: 10.1126/science.aac5560. [DOI] [PubMed] [Google Scholar]
- 30.Perkin MR, et al. Randomized Trial of Introduction of Allergenic Foods in Breast-Fed Infants. N Engl J Med. 2016;374(18):1733–1743. doi: 10.1056/NEJMoa1514210. [DOI] [PubMed] [Google Scholar]
- 31.Du Toit G, et al. Early consumption of peanuts in infancy is associated with a low prevalence of peanut allergy. J Allergy Clin Immunol. 2008;122(5):984–991. doi: 10.1016/j.jaci.2008.08.039. [DOI] [PubMed] [Google Scholar]
- 32.Du Toit G, et al. Randomized trial of peanut consumption in infants at risk for peanut allergy. N Engl J Med. 2015;372(9):803–813. doi: 10.1056/NEJMoa1414850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ierodiakonou D, et al. Timing of Allergenic Food Introduction to the Infant Diet and Risk of Allergic or Autoimmune Disease: A Systematic Review and Meta-analysis. JAMA. 2016;316(11):1181–1192. doi: 10.1001/jama.2016.12623. [DOI] [PubMed] [Google Scholar]
- 34.Katz Y, et al. Early exposure to cow’s milk protein is protective against IgE-mediated cow’s milk protein allergy. J Allergy Clin Immunol. 2010;126(1):77–82.e1. doi: 10.1016/j.jaci.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 35.Pitt TJ, et al. Reduced risk of peanut sensitization following exposure through breast-feeding and early peanut introduction. J Allergy Clin Immunol. 2018;141(2):620–625.e1. doi: 10.1016/j.jaci.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 36.Du Toit G, et al. Effect of Avoidance on Peanut Allergy after Early Peanut Consumption. N Engl J Med. 2016;374(15):1435–1443. doi: 10.1056/NEJMoa1514209. [DOI] [PubMed] [Google Scholar]
- 37.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469(7330):415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elder JB, Williams G, Lacey E, Gregory H. Cellular localisation of human urogastrone/epidermal growth factor. Nature. 1978;271(5644):466–467. doi: 10.1038/271466a0. [DOI] [PubMed] [Google Scholar]
- 39.Jaworek J, Konturek SJ. Distribution, release, and secretory activity of epidermal growth factor in the pancreas. Int J Pancreatol. 1990;6(3):189–205. doi: 10.1007/BF02924288. [DOI] [PubMed] [Google Scholar]
- 40.Knoop KA, et al. Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci Immunol. 2017;2(18):eaao1314. doi: 10.1126/sciimmunol.aao1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 2015;8(1):198–210. doi: 10.1038/mi.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kulkarni DH, et al. Goblet cell associated antigen passages are inhibited during Salmonella typhimurium infection to prevent pathogen dissemination and limit responses to dietary antigens. Mucosal Immunol. 2018;11(4):1103–1113. doi: 10.1038/s41385-018-0007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kulkarni DH, et al. Goblet cell associated antigen passages support the induction and maintenance of oral tolerance. Mucosal Immunol. 2020;13(2):271–282. doi: 10.1038/s41385-019-0240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobata R, et al. High levels of growth factors in human breast milk. Early Hum Dev. 2008;84(1):67–69. doi: 10.1016/j.earlhumdev.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Nojiri T, et al. Clinical significance of amphiregulin and epidermal growth factor in colostrum. Arch Gynecol Obstet. 2012;286(3):643–647. doi: 10.1007/s00404-012-2365-8. [DOI] [PubMed] [Google Scholar]
- 46.Sefik E, et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science. 2015;349(6251):993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solomon BD, Hsieh CS. Antigen-Specific Development of Mucosal Foxp3+RORγt+ T Cells from Regulatory T Cell Precursors. J Immunol. 2016;197(9):3512–3519. doi: 10.4049/jimmunol.1601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chai JN, et al. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol. 2017;2(13):eaal5068. doi: 10.1126/sciimmunol.aal5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453(7192):236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10(6):595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wohlfert EA, et al. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121(11):4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu M, et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 2018;554(7692):373–377. doi: 10.1038/nature25500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dallas DC, Underwood MA, Zivkovic AM, German JB. Digestion of Protein in Premature and Term Infants. J Nutr Disord Ther. 2012;2(3):112. doi: 10.4172/2161-0509.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mosconi E, et al. Breast milk immune complexes are potent inducers of oral tolerance in neonates and prevent asthma development. Mucosal Immunol. 2010;3(5):461–474. doi: 10.1038/mi.2010.23. [DOI] [PubMed] [Google Scholar]
- 55.Miyao T, et al. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36(2):262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 56.Rubtsov YP, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329(5999):1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10(9):1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lochner M, et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J Exp Med. 2008;205(6):1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abrams EM, Becker AB. Food introduction and allergy prevention in infants. CMAJ. 2015;187(17):1297–1301. doi: 10.1503/cmaj.150364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alvisi P, et al. Recommendations on complementary feeding for healthy, full-term infants. Ital J Pediatr. 2015;41:36. doi: 10.1186/s13052-015-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ladinsky MS, et al. Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science. 2019;363(6431):eaat4042. doi: 10.1126/science.aat4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Atarashi K, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163(2):367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hand TW, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337(6101):1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohashi Y, Hiraguchi M, Sunaba C, Tanaka C, Fujisawa T, Ushida K. Colonization of segmented filamentous bacteria and its interaction with the luminal IgA level in conventional mice. Anaerobe. 2010;16(5):543–546. doi: 10.1016/j.anaerobe.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 66.Jiang HQ, Bos NA, Cebra JJ. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect Immun. 2001;69(6):3611–3617. doi: 10.1128/IAI.69.6.3611-3617.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al Nabhani Z, Eberl G. Imprinting of the immune system by the microbiota early in life. Mucosal Immunol. 2020;13(2):183–189. doi: 10.1038/s41385-020-0257-y. [DOI] [PubMed] [Google Scholar]
- 68.Ramanan D, et al. An Immunologic Mode of Multigenerational Transmission Governs a Gut Treg Setpoint. Cell. 2020;181(6):1276–1290.e13. doi: 10.1016/j.cell.2020.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76(1):34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 70.Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132(7):2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 71.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci USA. 2005;102(14):5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eberl G, Littman DR. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004;305(5681):248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 74.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39(3):186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 75.McDonald KG, et al. Epithelial expression of the cytosolic retinoid chaperone cellular retinol binding protein II is essential for in vivo imprinting of local gut dendritic cells by lumenal retinoids. Am J Pathol. 2012;180(3):984–997. doi: 10.1016/j.ajpath.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McDole JR, et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483(7389):345–349. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lanik WE, et al. Breast Milk Enhances Growth of Enteroids: An Ex Vivo Model of Cell Proliferation. J Vis Exp. 2018;(132):56921. doi: 10.3791/56921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.