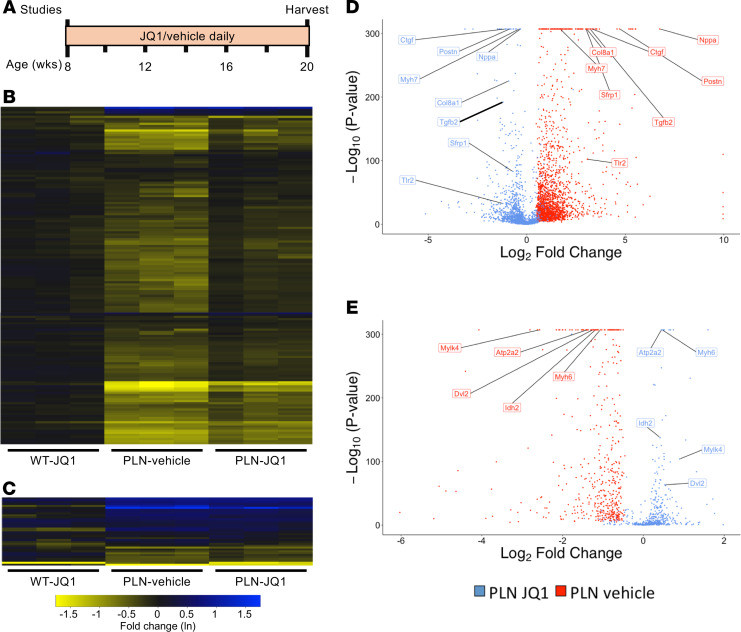
Figure 5. Chronic BET inhibition alters inflammatory gene expression in DCM.
(A) Experimental protocol (n = 3 mice/treatment group). Heat maps demonstrating that (B) genes controlling aerobic respiration (tricarboxylic acid cycle and mitochondrial oxidative phosphorylation) were almost uniformly downregulated, while (C) genes controlling cellular glucose utilization were generally upregulated in PLNR9C hearts. JQ1 had little effect on metabolic gene expression. Data plotted are natural log of fold-change values versus WT vehicle-treated mice. Volcano plots demonstrating the magnitude and significance of JQ1’s alteration of the expression of genes that were (D) upregulated or (E) downregulated in PLNR9C vehicle-treated hearts. In each plot, blue represents the effect of JQ1 on gene expression for all genes in red. Representative genes (labeled) reveal key processes affected by JQ1, including cardiac stress response signaling (Nppa, Myh6/Myh7), fibrosis and extracellular matrix remodeling (Postn, Col8a1), TGF-β signaling (Tgfb2, Ctgf), cytoskeletal signaling (Mylk4), Wnt signaling (Sfrp1, Dvl2), innate immune activation (Tlr2, Tlr4), and metabolism (Idh2, Atp2a2).