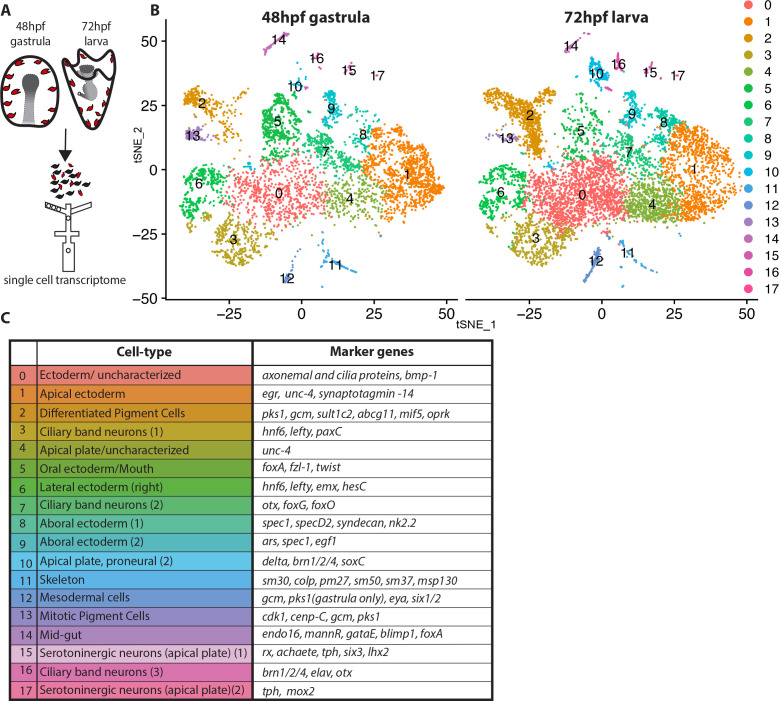
Figure 1. Dissection of S. purpuratus ectodermal cell-types by scRNA-seq.
(A) Cartoon summarizing the dissociation of gastrulae and larvae (pigment cells in red, other cell-types in black), cell separation through drop-seq technology and RNA sequencing. Our single cell sequence datasets detected 20,489 reads/genes and a median 2,909 UMI counts per cell at 48hpf as well as 19,716 reads/genes and a median 1,136 UMI counts per cell at 72hpf. (B) tSNE plots of gastrula (48hpf) and larvae (72hpf) enriched for the ectodermal cell-types. Colors indicate major cell-types grouped by gene expression similarity. In the 48hpf sample, 5688 single cells were captured for downstream analysis, sequenced at a level of 81,121 reads per cell. The 72hpf sample includes 8178 single cells with an average of 54,788 reads per cell. The two samples were integrated to identify conserved cell types and cluster marker genes using Seurat. (C) Table summarizing the cell-type for each cluster (48hpf +72hpf) with the most representative marker genes (colors in the table reflect colors of the tSNE plot clusters).