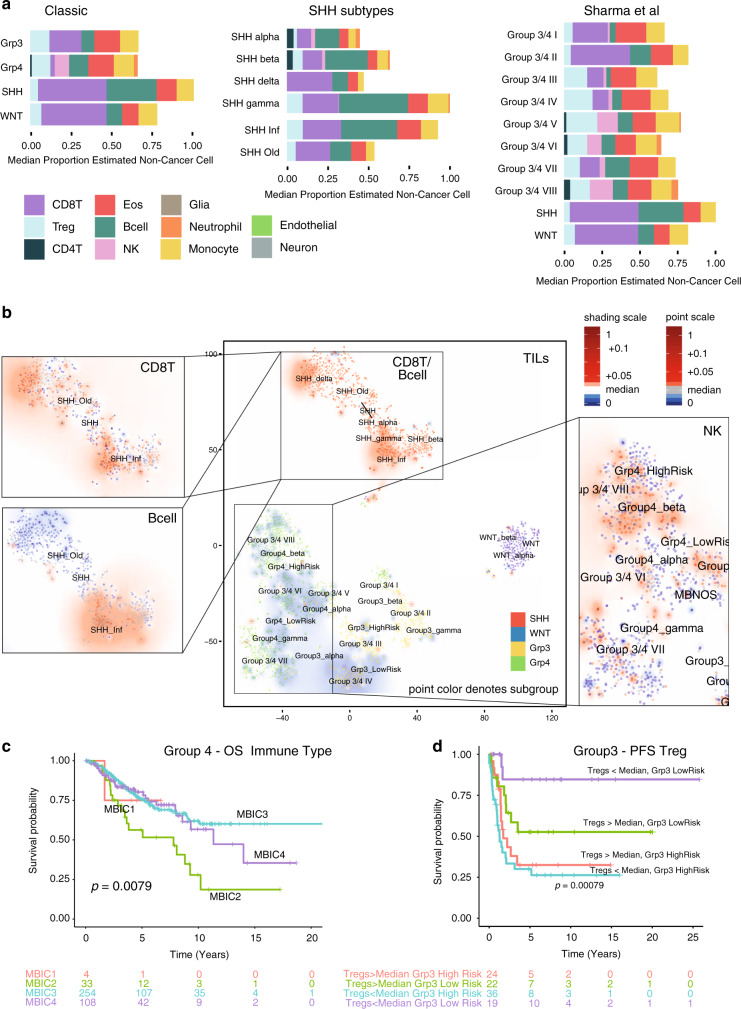
Fig. 3. Deconvolution analysis of medulloblastoma immune infiltration by subgroup.
a Barplots of the estimated median infiltration of specific cell types as a proportion of all non-cancer cell types (range scaled from 0 to 1) in 2325 medulloblastoma by subgroup (classic 4 medulloblastoma consensus subgroups42) by SHH subtype32,33 and by 10 group consensus as per Sharma et al.35. b t-SNE plot representing the methylation profiles of 2325 medulloblastoma. The colors of dots in the central panel map to the classic four molecular subgroups: red, SHH; blue, WNT; yellow, Grp3; green, Grp4. Text represents centroids of individual subtypes as reported variously by Cavalli et al.33, Sharma et al.35, and Schwalbe et al.32. Background shading represents the 2D spatial density estimation of the amount of tumor-infiltrating lymphocytes (TILs); red shading equals relatively greater than average infiltration and blue less than average. Exploded side panels represent enlarged areas of interest wherein both dot color and background shading represent the relative amount of the particular immune cell infiltration denoted. Red denotes relatively greater than average infiltration and blue less than average. c Kaplan–Meier plot showing significant difference in overall survival in MBGrp4 by immune cluster; log-rank p = 0.008, n = 399. d Kaplan–Meier plot showing significantly different progression-free survival (PFS) within the MBGrp3 Schwalbe et al.32 subtypes by low (<median) or high (>median) levels of Treg infiltration; log-rank p < 0.001, n = 101.