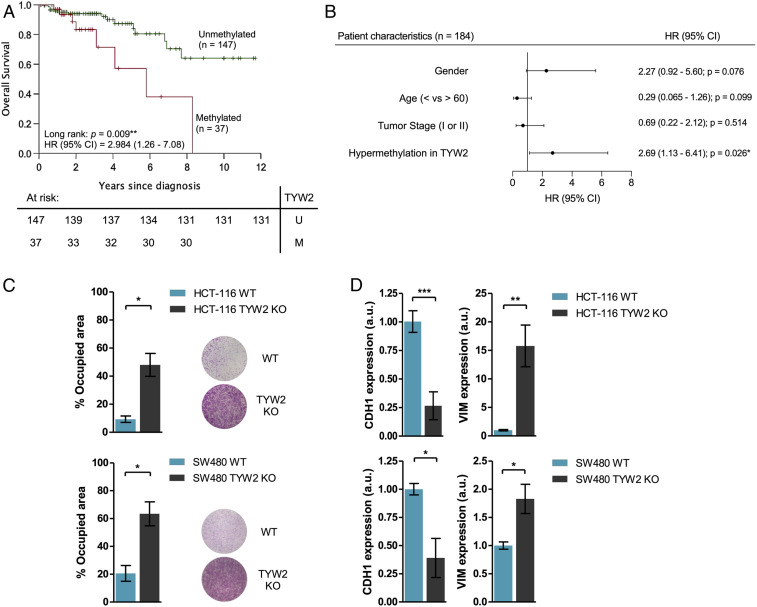
Fig. 4.
TYW2 epigenetic loss is associated with early-stage primary colorectal tumors with poor clinical outcome. (A) Kaplan–Meier curve showing that the presence of TYW2 hypermethylation in patients with early-stage colorectal cancer (n = 184) was significantly associated with shorter overall survival (P = 0.009). Green line, unmethylated cases; red line, methylated cases. **P < 0.01. (B) Forest plot representation of the Cox proportional hazard regression models demonstrating that TYW2 hypermethylation is an independent prognostic factor of poor overall survival in patients with early-stage colorectal cancer (HR = 2.69; 95% CI = 1.13 to 6.41; P = 0.026). ns, not significant; *P < 0.05; (C) Transwell assay to determine cell migration capability in the studied models. HCT-116 and SW480 cells carrying the CRISPR/Cas9 deletion of TYW2 showed a significant increase in the number of migrated cells after 48 h compared with wild-type cells. Representative images of the Transwell membrane staining are shown. P values are those associated with an unpaired two-tailed Student’s t test. *P < 0.05. (D) Assessment of EMT features in the studied models using qRT-PCR levels of the E-cadherin (epithelial) and vimentin (mesenchymal) markers. The KOs of TYW2 in the HCT-116 and SW480 cell lines showed a combined decrease in E-cadherin expression and enhancement Vimentin levels compared with wild-type cells, a transcript profile suggestive of the acquisition of EMT features. Data shown represent the mean ± SD of at least three biological replicates and were analyzed using an unpaired two-tailed Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.