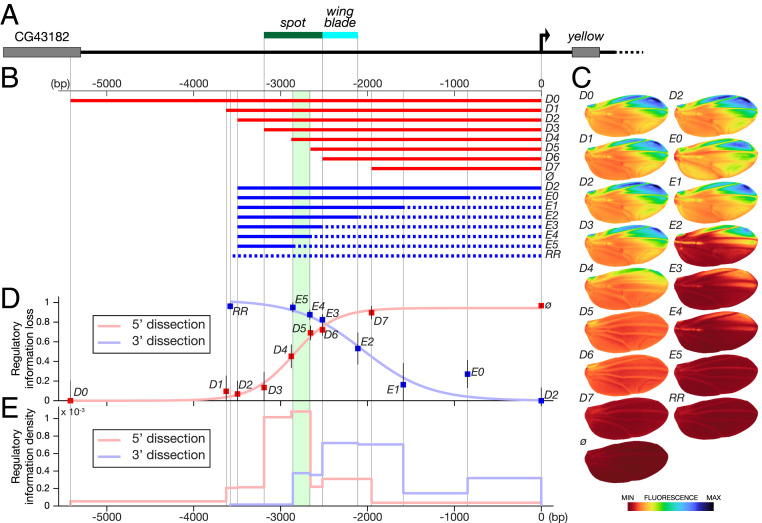
Fig. 1.
Quantitative mapping of wing regulatory activities at the yellow locus. (A) The top line represents the 5′ region of the yellow locus from D. biarmipes; the green and blue bars indicate the respective locations of spot and wing blade, respectively, as originally mapped (6). (B) Two series of fragments derived from y 5′ region (D series, red; E series, blue) were tested in reporter constructs in D. melanogaster. The dotted lines in the E series represent randomized sequences (Materials and Methods); ø and RR stand for an empty reporter vector and a vector containing a completely randomized fragment, respectively. The area shaded in green in B, D, and E identifies a previously studied regulatory component (16), spot196. (C) Images of average reporter expression of all individuals for each construct in the wing at emergence from the pupa according to the color map below. Note that spot196 appears strictly necessary to any activity in the spot region (compare D4 with D5 and compare E4 with E5). (D) Overall loss of regulatory information (fluorescence levels) along the sequence (base pairs). The loss of phenotypic information measures how much truncating or randomizing a fragment affects the whole activity relatively. It is estimated by the ratio where Px, Pref, and Pø are the phenotypes of construct x, construct D0 or D2 (the largest constructs of each series as a reference for that series), and the empty construct ø in the PCA space, respectively, plotted as a function of the distance to the starting point of the randomization (series E) or truncation (series D). Error bars represent the SD of the phenotype of each construct in PCA space normalized by the distance d(Pø,Pref). (E) Density of regulatory information along the y 5′ region (fluorescence levels per base pair). It is technically the first derivative of the regulatory information loss shown in D. For each series, it represents the phenotypic distance (in PCA space) between two consecutive constructs divided by the number of base pairs that changed between those two constructs. It indicates the regulatory contribution per base pair of each DNA segment measured in each series.