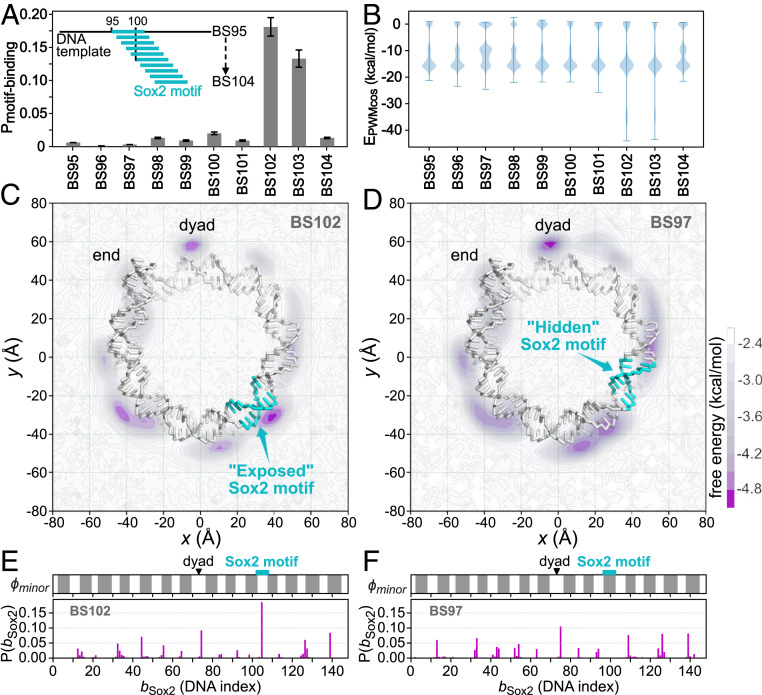
Fig. 3.
Sox2 binding to the modified 601 nucleosomes, in which the Sox2 consensus sequence “CTTTGTT” was inserted into the 601 sequence at different positions, with in the name BS meaning the insertion position. (A) Probability of Sox2 recognizing its consensus motif on the 10 modified 601 nucleosomes. Error bars are based on bootstrapping (SI Appendix, Supplementary Methods). (B) Distribution of the sequence-specific interaction energy between Sox2 and the nucleosomal DNA . (C) The 2D free energy surface of Sox2 center-of-mass (COM) on the BS102 nucleosome. DNA is shown as sticks, and histone proteins are hidden for clarity. The Sox2 motif is colored cyan. The purple intensity represents free energy value as depicted in the color bar. (D) The same as C but for Sox2 binding on the BS97 nucleosome. (E) One-dimensional distribution of the mean Sox2 binding positions on the BS102 nucleosomal DNA (, Lower) and the accessibility of the nucleosomal DNA minor groove (, Upper). The exposed regions that are accessible to Sox2 are shown in white , whereas the histone covered regions are shown in gray (see SI Appendix, Supplementary Methods for the detailed definition). The dyad and the Sox2 motif insertion positions are marked by a black triangle and a cyan rectangle, respectively. (F) The same as E but for the BS97.