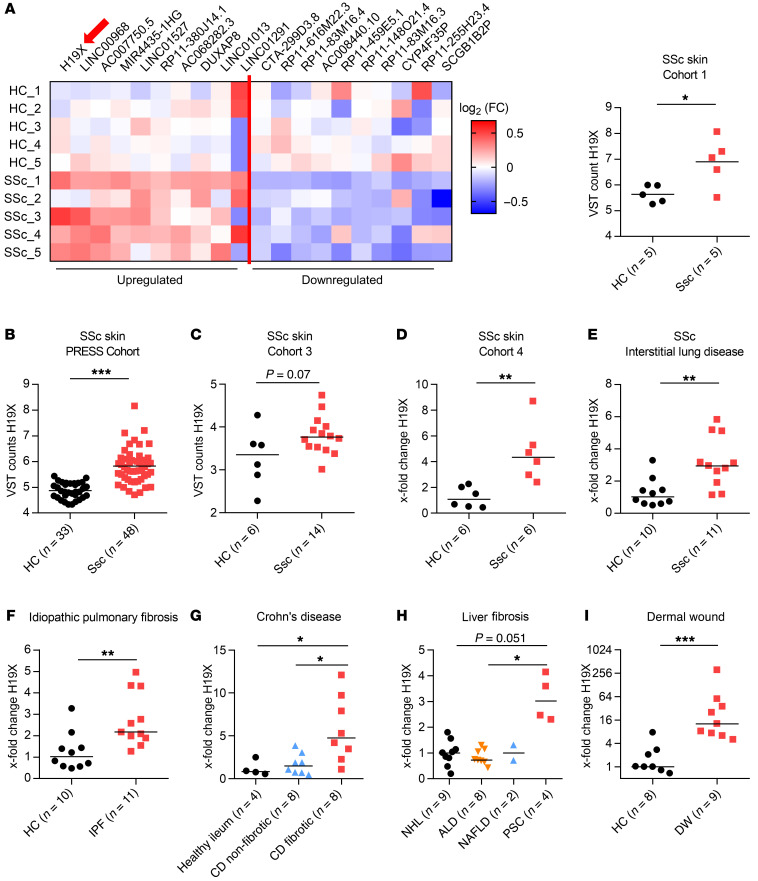
Figure 1. H19X is upregulated in skin from patients with SSc from different cohorts in a variety of fibrotic disorders and in physiological DW healing.
Heatmap showing 20 most significantly deregulated lncRNA in skin derived from patients with SSc as revealed by RNA-Seq analysis; H19X is indicated by the arrow (cohort 1) (A). H19X differential expression in SSc versus HC skin in the PRESS cohort (B), and cohort 3 (C), as measured by RNA-Seq. H19X differential expression as measured by qPCR in cohort 4, normalized to GAPDH and RPLP0 (D). H19X differential expression in ILD-SSc versus HC lung as measured by qPCR with normalization to GAPDH and RPLP0 (E). H19X differential expression in IPF (F), Crohn’s disease (G), PSC (H), and DWs measured by qPCR with normalization to GAPDH and RPLP0 (I). Data are presented as single values and median. Differential expression analysis was carried out on variance stabilized counts using DEseq2 package 44 (A–C). Mann-Whitney test (D–F and I). Kruskal–Wallis test (G and H). *P < 0.05, **P < 0.01, ***P < 0.001.