Abstract
Several cancer immunotherapy approaches, such as immune checkpoint blockade and adoptive T-cell therapy, boost T-cell activity against the tumor, but these strategies are not effective in the absence of T cells specific for displayed tumor antigens. Here we outline an immunotherapy in which endogenous T cells specific for a noncancer antigen are retargeted to attack tumors. The approach relies on the use of antibody- peptide epitope conjugates (APECs) to deliver suitable antigens to the tumor surface for presention by HLA-I. To retarget cytomegalovirus (CMV)-specific CD8+ T cells against tumors, we used APECs containing CMV-derived epitopes conjugated to tumor-targeting antibodies via metalloprotease-sensitive linkers. These APECs redirect pre-existing CMV immunity against tumor cells in vitro and in mouse cancer models. In vitro, APECs activated specifically CMV-reactive effector T cells whereas a bispecific T-cell engager activated both effector and regulatory T cells. Our approach may provide an effective alternative in cancers that are not amenable to checkpoint inhibitors or other immunotherapies.
Immunotherapies such as immune checkpoint blockade (ICB) have shown remarkable clinical benefit across a broad range of cancers1. However, the long-term success of this and other cancer immunotherapy approaches remains confined to a small minority of cancer patients. A mismatch between the specificity of T cells and the epitopes displayed on tumor human leukocyte antigen (HLA) plays a substantial role. A major determinant of response to PD-1 inhibitors is the tumor mutation burden (TMB)2,3, which contributes to the number of HLA-I-binding neoepitopes that can be recognized by CD8+ cytotoxic T lymphocytes (CTLs)4,5. Antigenicity is therefore rate limiting in nonresponding types of cancer characterized by low TMB, with the majority of cancers lacking sufficient neoantigens to achieve clinical benefit6,7.
The levels of CTLs are usually low in tumors, but their absence or paucity correlates with worse clinical outcomes in most cancer types8. Moreover, neoantigen-specific T cells are also typically found at low frequency within tumors9,10. In TMB-high tumors, vaccination efforts to expand CTLs against neoantigens have shown promising early results11–13. Others are developing methods to reprogram CTL specificity through genetic engineering approaches such as the introduction of T-cell receptors14 or chimeric antigen receptors15. These technologies correct the mismatch between antigens displayed on tumors and antigen-receptor specificity and have achieved promising clinical benefit16.
Here we sought to retarget virus-specific T cells against the tumor. In humans, immunity against persistent herpesviruses such as cytomegalovirus (CMV) is widespread in healthy individuals and often expands with age (a phenomenon termed ‘memory inflation’), giving rise to hyperexpanded clonotypes17,18. CMV has evolved sophisticated mechanisms to evade immunosurveillance for persistence in hosts, including many traits shared with tumors such as inhibition of HLA-I antigen processing and the inhibition of CTLs and natural killer cell function19. CMV-specific CTLs (CMV-CTLs) are present within tumors as ‘bystander-CTLs’ and, although they retain functional competence20,21, they are unable to target tumor cells due to the specificity of their T-cell receptors. We chose to retarget CMV-CTLs to tumors because of their high potency and abundance. To reprogram the antigenicity of tumor cells, we generated antibody-peptide epitope conjugates (APECs), which bind to target antigen on healthy and tumor cells and release immunogenic CMV epitopes after cleavage by tumor-specific proteases. The released peptide binds to free HLA class I molecules at the surface of the tumor cell and can be targeted for destruction by circulating CMV-CTLs.
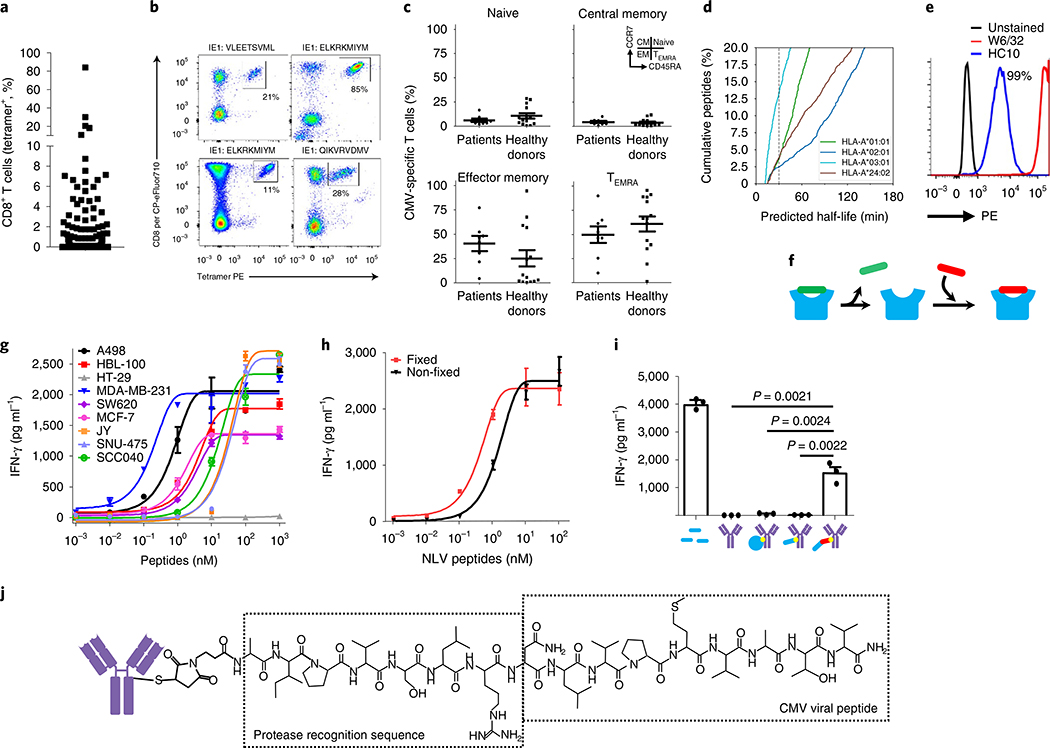
Previous studies have reported high frequencies of circulating CMV-CTLs in the peripheral blood of healthy individuals18. Using a panel of 11 HLA-peptide complexes covering known immunodominant CMV T-cell epitopes (Supplementary Table 1), we found high frequencies of circulating CMV-CTLs in cancer patients (Fig. 1a) with 33% having responses >0.5%. Some patients had exceptionally large responses, with up to 85% of all CD8+ T cells being specific for a single CMV peptide antigen (Fig. 1b). Comparing CMV-CTLs in patients and healthy donors there is no significant difference in the phenotype of CMV-CTLs, with the majority of cells being either effector memory (TEM) or terminally differentiated effector mem- ory-revertant cells (TEMRA) (Fig. 1c), suggesting that this immunity is functionally intact.
Fig. 1 |. CMV-CTLs are found at high frequencies in patients with cancer and recognize antigen-modified cancer cells.
a, CMV-specific immunity in healthy donors (n = 30) and cancer patients (n = 104) is found at high frequencies. b, Example tetramer staining. Percentages denote tetramer+ cells of the total CD8 T-cell population. PE, phycoerythrin. c, Memory phenotype analysis of CMV-CTL populations between healthy donors (n = 14) and cancer patients (n = 8) using CCR7 and CD45RA expression. d, In silico prediction of dissociation rates for HLA-bound peptides from surface. e, Flow cytometric analysis of peptide-loaded (W6/32) and empty HLA molecules (HC10) at the cancer cell line surface. Percentages denote cells positive for HC10 staining. All cell lines tested were 100% positive for peptide-loaded HLA (W6/32). Staining was repeated (n = 3) in selected cell lines. f, Schematic of peptide dissociation and peptide loading into HLA at the cell surface. g, IFN-γ release depicting T-cell activation in response to eight tumor cell lines with exogenously added CMV peptide (n = 3 independent samples). h, T-cell cytokine release after recognition of peptide-loaded tumor cells that were loaded in either a fixed or unfixed state (n = 3 independent samples). i, T-cell recognition of tumor target cells pretreated with either exogenously added free CMV peptide (1 μM) or antibody conjugates (350 nM) where the conjugate was either no payload, whole-protein antigen, minimal CMV epitope peptide or protease-cleavable CMV peptide (n = 3 independent samples). g-i, Data shown as mean and error bars represent s.e.m. Significance was determined by unpaired two-tailed f-test. j, Chemistry schematic of APECs. In relevant panels, error bars represent s.e.m.
HLA-peptide complexes are noncovalently linked heterotrimers composed of the HLA heavy chain, a peptide antigen and a β2-microglobulin molecule. The affinity of bound peptides for HLA molecules is known to span a wide range. Using in silico prediction methods, we calculated the half-life and affinity from 8,211 previously identified peptides eluted from surface HLA proteins22. From this analysis an estimated 2.5–12.5% of peptides have a half-life < 30 min (Fig. 1d) and 20–50% have a low predicted binding affinity >100 nM (Supplementary Fig. 1). We hypothesized that a sizable fraction of HLA molecules would therefore be empty at the surface of cancer cells. To test this, we used the monoclonal antibody W6/32, which is reactive toward properly folded peptide-bound HLA molecules, and HC10, which is reactive toward empty/unfolded HLA-B and HLA-C molecules23. This revealed strong staining of HC10 in all cell lines, confirming the presence of empty HLA molecules at the surface of tumor cells cultured in vitro (Fig. 1e and Supplementary Fig. 2a).
We reasoned that empty HLA molecules may be permissive to surface loading with exogenously provided peptide (Fig. 1f). Indeed, using a panel of eight HLA-A*0201 (A2) cell lines we could passively load the CMV immunodominant peptide antigen NLVPMVATV (NLV) efficiently in each case such that CMV-CTLs could recognize and kill cells exposed to nanomolar levels of free peptide (Fig. 1g and Supplementary Fig. 2b). To address whether peptides were being loaded into HLA molecules at the cell surface or following cellular uptake, we repeated the experiment using lightly fixed tumor cells that are unable to take up exogenous peptide. T-cell recognition was almost identical between fixed and non-fixed target cells, suggesting that peptides were loaded directly into empty HLA molecules at the cell surface (Fig. 1h). These data demonstrate that the HLA-peptidome of tumor cells can be efficiently manipulated through the provision of exogenous peptide antigen at the cell surface, potentially creating an opportunity to redirect pre-existing tumor-resident cytotoxic immunity against tumors through antigenic reprogramming.
Antibodies have been used successfully to deliver cytotoxic payloads to tumors for a range of malignancies and are the obvious vector for delivery of peptide antigens. We tested the ability of antibodies to deliver antigens to the surface of tumor cells by conjugating antibodies (Ab) with CMV whole-protein antigen (pp65) or the CMV peptide epitope (NLV). Neither Ab-protein nor Ab-peptide was able to reprogram surface antigenicity, yet peptides that had been modified to contain protease cleavage sites for the cancer-associated protease matrix metalloprotease 2 (MMP2) were able to activate CMV-CTLs (Fig. 1i; P = 0.0022 Ab-protease versus Ab-peptide; P = 0.0024 Ab-protease versus Ab-protein; P=0.0021 Ab-protease versus Ab; unpaired one-sided f-test). These data suggest that antibodies bound to engineered peptide antigens can efficiently reprogram surface antigenicity. Conjugation of peptides, assayed by high-pressure liquid chromatography (HPLC), demonstrated an average conjugation ratio of two peptides per antibody (minimum of zero peptides and maximum of eight; Supplementary Fig. 3). We termed these molecules APECs (Fig. 1j).
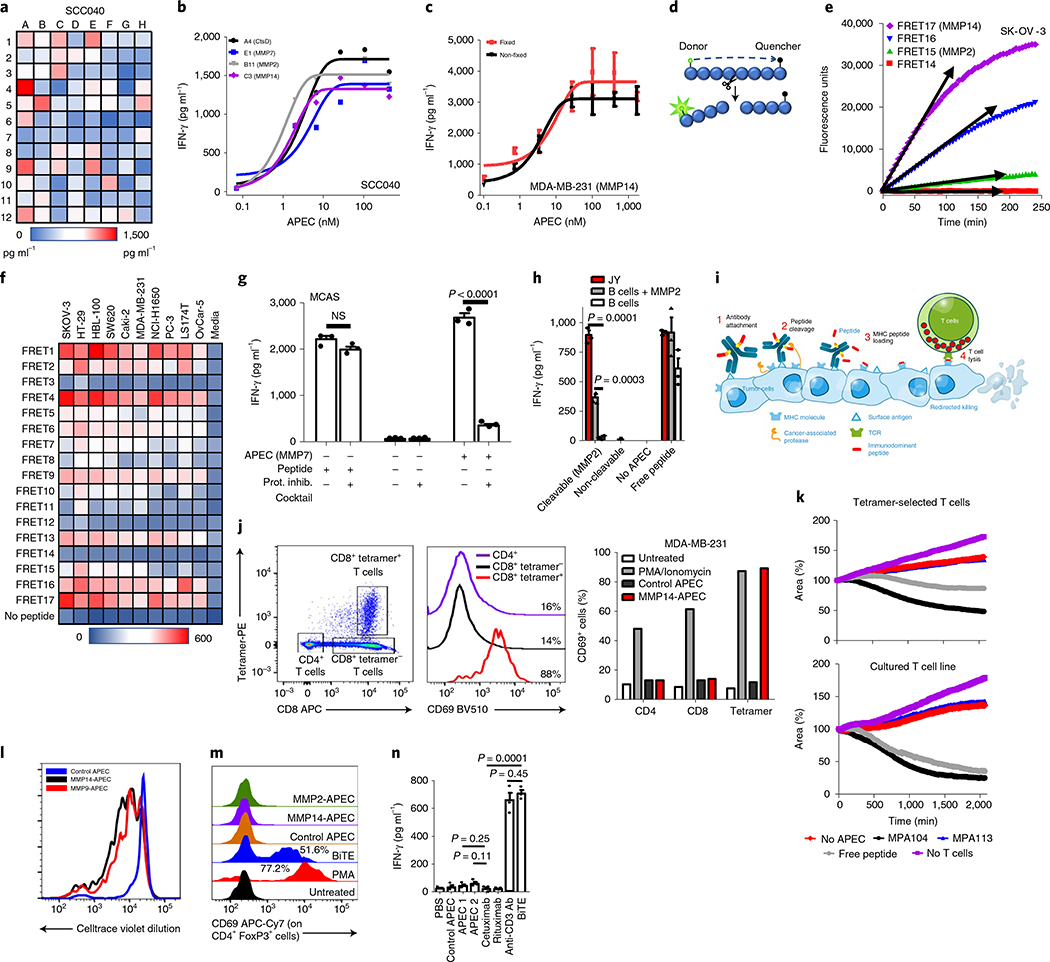
Tumors are known to express a wide range of proteases that have been implicated in processes such as tissue invasion and metastasis24. These have been exploited to release cytotoxic payloads and to increase target specificity for therapeutic antibodies25. To identify optimal protease sites, a library of 96 APECs was created based on the clinically approved anti-epidermal growth factor receptor (EGFR) antibody cetuximab (cAPEC) that contained short protease recognition sequences for cancer-associated proteases (Supplementary Table 2). This was used to identify peptides that optimally engaged CMV-CTLs against nine cancer cell lines (Fig. 2a and Supplementary Fig. 4), and the potency of lead APECs representing four different proteases was then determined (Fig. 2b). Further, multiple ex vivo expanded CMV-specific T-cell lines were used with peptide specificity ranging between 39 and 96% (Supplementary Fig. 5). There was no difference in the efficacy of APEC when using T-cell lines with higher or lower frequency of peptide-specific T cells. We used real-time imaging to examine the kinetics of MDA-MB-231 killing by CMV-CTLs in vitro using MMP2-cAPEC, which revealed progressive killing over 48 h (Supplementary Videos 1–5).
Fig. 2 |. Engineering APECs to deliver peptides to the surface of tumor cells.
a, Tumor cell line treated with 96different cAPECs screened using a functional CMV-CTL cytokine release assay. b, APECs demonstrating positive responses in a were used in a dose-response assay to determine median effective concentration required to induce a 50% effect. CtsD, cathepsin D. c, Functional T-cell cytokine release assay demonstrating recognition of unfixed and lightly fixed cells treated with cAPEC (n = 3 independent samples). d, Schematic depicting FRET assay. e,f, Example data of peptide cleavage kinetics of FRET assay (maximum voltage depicted by arrows) (e) and with data from ten tumor cell lines shown (f). g, Antigenic reprogramming of tumor cells by APEC was inhibited by the co-incubation of protease inhibitors (n = 3 independent samples). Significance was determined by unpaired two-tailed f-test. h, MMP2 rAPEC-treated healthy B cells (350 nM) did not activate T cells whereas malignant B cells (JY) efficiently activated them. Healthy B cells become susceptible to APECs following the provision of exogenous MMP2 protease. Addition of exogenous free CMV peptide (1 μM) allowed recognition of both healthy and tumor B cells (n = 3 independent samples). Significance was determined by unpaired two-tailed f-test. i, Schematic of APEC showing the proposed mechanism of action at the tumor cell surface. j-l, APEC-treated tumor cells activated only the peptide antigen-specific T-cell population within PBMCs, as depicted by tetramer and CD69 flow cytometric analysis (j). Percentages denote activated T cells compared to untreated control (data from a single experiment). Comparison of cytotoxicity of CMV-CTL taken directly from PBMCs or in vitro expanded against APEC-treated MDA-MB-231 tumor cells (k); data from a single experiment. Proliferation of CellTrace violet-labeled CMV-CTLs isolated from PBMCs against APEC-treated MDA-MB-231 cells (l); data from a single experiment. m, Regulatory T cells were activated by BiTE but not by any of the APECs tested, as identified by flow cytometric analysis using the T-cell activation marker CD69. Percentages denote activated Treg compared to untreated control (data from a single experiment). n, Immobilized BiTE and the anti-CD3 antibody OKT3 activated T cells whereas no such activation was observed using immobilized APEC (n = 3 independent samples). Significance was determined by unpaired two-tailed f-test. c,g,h,n, Data shown as mean and error bars represent s.e.m.
To demonstrate that peptide epitopes were being loaded at the cell surface we repeated the experiment on lightly fixed target cells, revealing similar targeting efficiency (Fig. 2c). Using HC10 antibody staining, we demonstrated a modest decrease in HC10 staining intensity after APEC labeling, which suggests peptides from APEC being loaded into empty HLA-I molecules at the surface of tumor cells (Supplementary Fig. 6). To determine the kinetics of peptide cleavage at the cell surface, we designed a Forster resonance energy transfer (FRET)-based peptide assay to identify the real-time cleavage of peptide substrates by tumor cells (Fig. 2d and Supplementary Table 3). These data showed high variability and, in many cases, rapid cleavage of peptides (Fig. 2e), which we ran against a panel of ten cell lines (Fig. 2f). This assay allowed us to identify sequences that were frequently cleaved by proteases active on many tumor cell types (for example, MMP2, MMP9, MMP14 and cathepsin B and D).
To confirm the mechanisms by which APECs induced antigenic reprogramming, we used protease inhibitors and recombinant proteases to verify the dependency on cellular protease activity. As expected, protease inhibitors substantially reduced the activation of T cells when added with APECs to tumor cells yet had no effect on free peptide incubation (Fig. 2g) (P < 0.0001, unpaired t-test).
The therapeutic promise of APECs is dependent on the ability to selectively reprogram the antigenicity of tumor but not of normal tissue. To explore the effect of APECs in reprogramming healthy tissue, we developed APECs based on the anti-CD20 therapeutic rituximab. Rituximab-APECs (rAPECs) were developed that actively reprogrammed the surface of lymphoma cell lines using the MMP2-cleavable sequence. The malignant B cell line JY was efficiently antigenically reprogrammed using rAPECs yet healthy B cells were not (Fig. 2h) (P=0.0001, unpaired one-sided f-test). Importantly, the addition of exogenous MMP2 protease was able to render healthy B cells susceptible to antigenic reprogramming (Fig. 2h) (P = 0.0003, unpaired one-sided t-test).
To confirm that the antibody specificity of APECs is critical for target activity, we used cAPECs and rAPECs on antigen-expressing and non-expressing cell lines. In all cases, APECs were able to reprogram target antigen-expressing tumors at lower concentrations than antigen-negative cells (Supplementary Fig. 7). Overall, these data support the mechanism of antigenic programming of tumor cells using APECs as requiring the binding of antibody to the surface of the target cell, the protease-dependent release of peptide and the loading of empty HLA molecules at the cell surface (Fig. 2i).
A major limitation of clinically approved T-cell engagement of therapeutic antibodies such as BiTEs relates to their nonspecific T-cell activation that is associated with high morbidity and mortality26. BiTE agents trigger T-cell activation through CD3ε, which results in them becoming activated and probably contributing to cytokine release syndrome seen in patients. In theory, limiting of T-cell activation to defined antigen-specific CTL populations may decrease the risk or severity of this syndrome.
To date, approaches that redirect T cells against cancer have shown clinical efficacy only in liquid tumors. This may, in part, be due to the high proportion of regulatory T cells (Treg cells) found in solid tumors that may also become activated and oppose anti-tumor CTLs27,28. APECs may obviate both of these problems by limiting T-cell activation to a subset of CTLs, reducing the proportion of T cells that becomes activated and avoiding the activation of undesirable immune cells such as Treg cells.
To test this, we used subpopulation analysis of bulk circulating blood cells to define the proportion and characteristics of immune cells activated by APECs. We designed an MMP14-activated cAPEC (peptide sequence: PRSAKELRNLVPMVATV) and an inactive control cAPEC that lacked a protease recognition site (peptide sequence: GGGSGGGSNLVPMVATV) and compared immune target specificity to the CD19-specific BiTE blinatumomab. We used peripheral blood mononuclear cells (PBMCs) because they contain a diverse and broad range of immune effector cells that allowed us to pinpoint APEC-activated cells.
Experiments using APECs revealed that only epitope-specific CTL were activated by MMP14-cAPEC, with no other T-cell populations showing evidence of stimulation (Fig. 2j). Furthermore, epitope-specific CTL freshly isolated from PBMCs demonstrated cytotoxic capacity (Fig. 2k) and proliferative capability (Fig. 2l and Supplementary Fig. 8a) in response to APEC-labeled tumor cells. To examine the differential activity of APECs and BiTEs on CD4+CD45RA+FoxP3+Treg cell activation (Supplementary Fig. 8b), we designed MMP2, MMP14 and control rAPECs against target healthy B cells within PBMCs. BiTEs induced strong activation of Treg cells whereas this was not observed with any of the APECs (Fig. 2m).
Additionally, immobilized APECs were unable to activate T cells, which contrasts with immobilized BiTEs and anti-CD3-specific antibodies that potently activate them (Fig. 2n; P = 0.0001 BiTE versus cetuximab, P = 0.0003 anti-CD3 versus cetuximab; unpaired one-sided t-test).
A potential constraint of antigenic reprogramming relates to the HLA restriction of each epitope and the variability of immunity against chronic viral infection. This could be a limitation for deployment of APECs in the clinic for diverse patient populations. Therefore, to move beyond a single HLA allele, we developed cAPECs that were conjugated to non-HLA-A2-restricted peptides. Using the same MMP2 protease cleavage sequence, cAPECs containing CMV epitopes presented by HLA-A*01:01 (A1) and HLA- B*08:01 (B8) demonstrated antigen-specific T-cell responses similar to those seen with the A2-restricted peptide (Supplementary Fig. 9). An advancement of this concept relates to the development of APECs containing multiple payload species that would both expand the number of T cells activated and also facilitate the development of APECs that could be deployed across HLA barriers. We tested three different ‘polytope’ APECs that contained either concatemers, mixed peptides or branched peptides (Supplementary Fig. 10). Each of these formats was able to activate multiple T-cell populations, providing at least proof-of-concept data that APEC technology can be used to deliver multiple T-cell peptide antigens to tumors.
Furthermore, we could expand the antiviral T-cell responses utilized by APECs to include immunodominant peptides from other herpesviruses including Epstein-Barr virus (EBV). We generated rAPECs using the A2-restricted EBV-derived epitope GLCTLVAML and, using the JY cell line, demonstrated T-cell activation/recogni- tion of rAPEC-labeled cells (Supplementary Fig. 11a; P = 0.0059, unpaired one-sided f-test). Finally, we demonstrate that we can use cAPECs with different protease cleavage sequences to target different tumor types. We created cAPECs containing MMP9, MMP2, ADAM28 and MMP14 sequences that were able to target multiple tumor types including lung and breast cancer cell lines (Supplementary Fig. 11b).
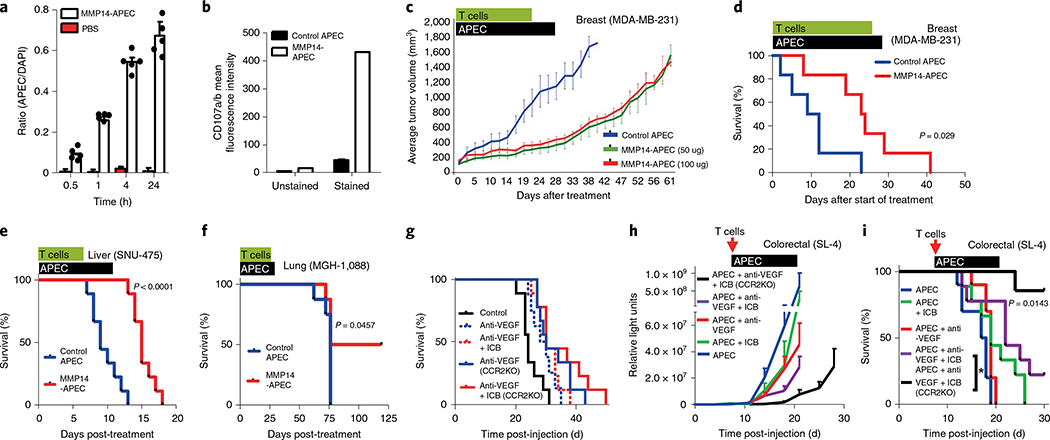
After demonstrating the efficacy of APEC in vitro, we moved forward to determine whether we could reprogram the antigenicity of tumors in vivo. First, we demonstrated that APEC can penetrate the center of the tumor (Fig. 3a and Supplementary Fig. 12) as well as its properties of activation (Supplementary Fig. 13a,b) and cytotoxic capability, using CD107 staining (Fig. 3b) of CMV-CTLs in vivo. We chose to test the broadly reactive MMP14-cAPEC against three different human tumors (breast, liver and lung) growing as established xenografts in immunodeficient mice reconstituted with adoptively transferred human CMV-CTLs. For the lung cancer model, we used a patient-derived xenograft (PDX) model MGH- 1088 (ref. 29). In each case, MMP14-APEC was able to delay the outgrowth or extend the survival of mice compared to the control APEC (Fig. 3c–f). The increased efficacy of APECs in the hepatocellular carcinoma model (Fig. 3e) may reflect the increased vascularity of liver tumors, and remains an area of interest. We were also able to demonstrate the presence of resident peptide-specific T cells 7 d after intravenous injection, using HLA-peptide tetramer staining within tumors from 5/5 mice with lung cancer after excision and dissociation of the tumor (Supplementary Fig. 13c,d).
Fig. 3 |. APEC modulation of antigenicity in vivo.
a, In vivo tumor penetration of APEC-treated NOD/SCID mouse model of primary orthotopic human breast cancer (MDA-MB-231) using the ratio of APEC to DAPI (n = 4 independent samples). Data shown as mean and error bars represent s.d. b, In vivo T-cell cytotoxic capacity, as assayed by CD107 staining, demonstrated increased cytotoxic activity of freshly isolated CMV-CTL in MDA-MB-231 treated with MMP14-cAPEC compared with control cAPEC. c, NOD/SCID mouse model MDA-MB-231 (n = 8) demonstrated a delay in tumor growth following treatment with 50 μg or 100 μg MMP14 cAPEC compared with control cAPEC. d, MMP14-cAPEC improved the survival of mice (n = 8) in a breast cancer neoadjuvant model compared with control cAPEC (P = 0.029, Mantel-Cox two-sided test) after injection of ex vivo expanded CMV-CTL. e, Survival of mice orthotopically injected with hepatocellular carcinoma cell line SNU-475 (n = 8) treated with MMP14-cAPEC and ex vivo expanded CMV-CTL was improved compared to that of mice treated with control cAPEC (P< 0.0001, Mantel-Cox two-sided test). f, Improved survival of mice (n = 5) treated subcutaneously with MMP14 or control cAPEC and ex vivo expanded CMV-CTL in a PDX model of lung cancer (P = 0.0457, Mantel-Cox two-sided test). g, C57BL/6 or Ccr2−/− (C57 background) mouse model (n = 10) of metastatic colorectal carcinoma (SL4 clone) demonstrated only a marginal increase in survival when treated with either anti-VEGF or anti-VEGF in combination with ICB therapy (anti-PD-1+ anti-CTLA-4). All mice received ex vivo expanded OT-I T cells specific for SIINFEKL. h,i, Ccr2−/− mice (n = 8–10) treated with combination immune checkpoint blockade (therapy of anti-VEGF+ ICB+ murine APEC system demonstrated significant delay in tumor growth (h) and increased survival (i) compared with wild-type mice treated with the same reagents or with other APEC combination therapies (P = 0.0143, Mantel-Cox two-sided test). All mice received ex vivo expanded OT-I T cells specific for SIINFEKL. In relevant panels, error bars represent s.e.m.
Finally we tested the efficacy of APEC using murine tumor models in syngeneic immunocompetent mice. To achieve this, we first developed anti-EGFR APEC based on the anti-murine EGFR antibody D1D4J by screening peptides containing the OT-1-specific antigen SIINFEKL (Supplementary Fig. 14a,c and Supplementary Table 4). OT-1 cells were primed ex vivo by peptide activation before adoptive transfer into recipient immunocompetent mice. Subcutaneous tumor models typically used for immunotherapy experiments are responsive and are thus considered not to align closely with the clinical disease. We found that murine liver metastatic colorectal cancers (mCRCs) are refractory to standard of care such as anti-vascular endothelial growth factor (VEGF) and chemotherapy30, as well as ICB (Fig. 3g). Similarly, APEC as a single agent was unable to control the outgrowth of SL4 tumors (Supplementary Fig. 14d) using ex vivo expanded OT-1 T cells specific for SIINFEKL. Although the combination of APEC plus ICB somewhat improved liver mCRC response, the effect was modest (Fig. 3h,i). We have previously shown elevated myeloid cells in liver mCRCs, especially with anti-VEGF treatment30. We therefore hypothesized that targeting of immunosuppressive myeloid suppressor cells (MDSCs) in the tumor microenvironment could enhance response to immunotherapy in liver mCRCs. While the depletion of monocytic MDSCs by the CCR2 gene KO only marginally improved ICB with anti-VEGF therapy for mCRCs (Fig. 3g), APEC therapy markedly improved the survival of mice with these combined treatments (Fig. 3h,i; P = 0.0143, Mantel-Cox two-sided test). These data show that APECs are able to redirect nonviral- specific T-cell responses against tumors, especially in combination with the reprogramming of the immune tumor microenvironment such as MDSC depletion.
Recent data have emerged demonstrating the ability of antigenic reprogramming by the injection of viral peptides directly into tumors in mice31. This work demonstrates the ability of viral T cells to control tumors in vivo, particularly when used in combination with anti-PD-1 therapies.
Our results describe the basis for engineering tumor antigenicity using proteolytic release of immunodominant viral epitopes from therapeutic antibodies. We anticipate that technologies such as APEC may yield new therapeutic opportunities for patients with cancer who have intact antiviral immunity. Ultimately, such technologies may need to be designed in a more personalized manner than other mainstream immune-oncology agents, but would potentially achieve a more targeted effect. This may include the need to profile the proteolytic nature of tumors and the composition of viral-specific T cells. The feasibility of this approach may be supported by delivering multiple peptide payloads that target a broader population of tumor cells and increase patient coverage using a single targeting entity.
Overall, our study provides an avenue for redirection of bystander T cells to the tumor, enabling an immunotherapy that could be delivered to low-TMB tumor types where there is considerable unmet clinical need.
Methods
Cell lines
All human tumor cell lines were purchased from ATCC. The SL4 mouse CRC cells were a generous gift from Professor Irimura at Tokyo University. Firefly luciferase+ EBV-transformed B cell lines (FFLuc) were a kind gift from B. Savoldo (UNC-Chapel Hill, USA). All cell lines were grown according to ATCC directions. Tumor cell lines were cultured in either complete Dulbecco’s modified Eagle’s medium (DMEM, Corning), DMEM/F-12 (Thermo Fisher) or RPMI (Corning), supplemented with 10% heat-inactivated fetal bovine serum (FBS, Thermo Fisher), 100 U ml−1 each of penicillin and streptomycin (Thermo Fisher) and 4 mM of L-glutamine (Life Technologies)). B cell lines (FFLuc and JY) were cultured in complete RPMI medium (Corning). All cell lines and assay cultures were maintained at 37 °C and 5% CO2. All cells were tested regularly for both mycoplasma contamination and rodent pathogens; no cell lines tested positive at any point.
In vitro expansion of peptide-specific T-cell lines
Peripheral blood mononuclear cells from healthy donors were used to culture peptide-specific T-cell lines. Briefly, 10 × 106 PBMCs (5 × 106 ml−1) were stimulated with 10 ng ml−1 of peptide in RPMI medium supplemented with 10% heat-inactivated FBS (Thermo Fisher), 100 U ml−1 each of penicillin and streptomycin (Thermo Fisher), 4 mM of L-glutamine (Life Technologies) and 1% human AB serum (Sigma). Following culture of cells for 4 d at 37 °C, 50% of the media was removed and replaced with the same media supplemented with 500 IU of interleukin-2 (IL-2, Peprotech). The cells were fed every 2–3 d and expanded for up to 6 weeks. On day 21, T cells were stained with HLA-peptide tetrameric complex for flow cytometric analysis and only those cell lines with >50% peptide-specific T cells were used in functional assays.
Peptides
Peptides used in this study were synthesized with Fmoc chemistry, isolated by HPLC to >90% purity and validated with mass spectrometry (Genscript). Further modifications were made to contain an N-terminal 3-maleimido propionic acid (3-MPA) group to allow chemical conjugation to free thiol groups and a C-terminal amide group. CMV-derived epitopes incorporated into the peptide sequences were NLV495–502 (NLVPMVATV), TPR417–476 (TPRVTGGGAM), YSE363–373 (YSEHPTFTSQY), VTE245–253 (VTEHDTLLY), ELK199–207 (ELKRKMIYM), GLC259–267 (GLCTLVAML) and CLG426–434 (CLGGLLTMV). Peptides were dissolved to 10 mM in DMSO for conjugation.
Conjugation of peptide to antibody
The antibody was buffered using 5% v/v 1 M Tris/25 mM EDTA pH 8.0. Disulfide bonds within the antibody were reduced by equimolar addition of 10 mM Tris(2-carboxyethyl)phosphine (TCEP) for 90 min. Peptides containing an N-terminal 3-maleimido propionic acid (3-MPA) group were added to the reduced antibody at 3× molarity of TCEP for 60 min before the addition of 10 mM N-acetyl cysteine (at 3 × molarity of TCEP) to quench free, unbound peptide. Antibody-peptide complexes were then purified from unbound peptide either by protein A purification (GE Healthcare) or ultrafiltration (Millipore). Complexes purified using protein A were eluted from protein A sepharose beads using 0.1 M glycine (pH 2.7) and buffered with 1 M Tris (pH 8.0). Complexes purified using ultrafiltration were diluted to 500 μl in PBS and concentrated to 25 μl. This was repeated a total of eight times before dilution to 250 μl in PBS. Protein quantitation utilized absorbance readings at 280 nm using a NanoDrop 2000 (Thermo Fisher). Purified APECs were diluted to required concentrations using PBS and stored at 4 °C for up to 2 weeks.
Blood sampling
Deidentified human blood was obtained from healthy adult donors under informed consent (Massachusetts General Hospital, Blood Transfusion Service). Deidentified human blood from patients with cancer (lung, head and neck, breast cancer) was collected from patients undergoing routine follow-up. All protocols were approved by the Massachusetts General Hospital Institutional Review Board and performed in accordance with Federal Law. The study was conducted according to the provisions of the Declaration of Helsinki, and all patients signed and provided their informed consent before sample collection.
Peripheral blood mononuclear cells were isolated using Ficoll-Paque Plus (GE Healthcare) density gradient centrifugation according to the manufacturer’s instructions. For long-term storage, PBMCs were resuspended in FBS with 10% DMSO and stored in liquid nitrogen at a density of 1–5 × 107 cells ml−1.
Flow cytometry
Cells were washed in PBS+ 2% serum and placed in 5-ml FACS tubes (Corning Falcon). Antibodies (BioLegend) were added at 1/100 dilution and HLA-peptide tetramers containing CMV epitopes (National Institute of Health Tetramer Core Facility, Emory University, USA) were added at 1/200. Cells were incubated for 30 min at room temperature in the dark, and excess antibody/ tetramer washed off using PBS+ 2% serum. Where necessary, a secondary anti-mouse IgG PE (Thermo Fisher) was added to the cells and excess antibody removed by washing in PBS+ 2% serum. Labeled cells were analyzed using the Fortessa X-20 (Becton Dickinson). Flow cytometry data were analyzed using FlowJo X software (FlowJo).
In vitro functional T-cell assay to determine efficacy
Target cells were labeled with APECs (0.05–300 nM) in the serum-free medium Aim-V (Thermo Fisher) for 30 min at room temperature, and excess APECs removed by washing in fresh media. Labeled target cells were incubated overnight in the presence of peptide- specific T cells in serum-free media at an effector-to-target (E/T) ratio of 1/10, and supernatant assayed for T-cell stimulation or at an E/T ratio of 1/1 and the supernatant assayed for cytotoxicity after 16–20 h. Supernatant was used to detect T-cell activation by the presence of interferon (IFN)-gamma release by ELISA, with the Ready-Set-Go Human IFN gamma ELISA kit (eBioscience), or by T-cell cytotoxicity using the CytoTox assay kit (Promega) to assay for the presence of lactate dehydrogenase released by dead/dying target cells.
T-cell proliferation using dilution of CellTrace violet
T cells were loaded with CellTrace violet (Thermo Fisher) according to the manufacturer’s protocol. Briefly, T cells were washed in PBS, resuspended to 1 × 106 ml−1 in PBS and 1 μl of CellTrace violet (10 μM) per 1 ml of T cells and incubated at 37 °C for 20 min. Five volumes of PBS+ 2% serum was added to cells and incubated at 37 °C for 5 min. Cells were pelleted by centrifugation, resuspended in RPMI+ 10% FBS and incubated at room temperature for 15 min. Cells were pelleted and resuspended at appropriate dilution for use in the assay.
T-cell cytotoxicity using time-lapse microscopy
Tumor cells were plated in 96-well flat-bottom plates and left to adhere overnight at 37 °C. T-cell functional assays were set up as previously described, with excess APECs removed before the addition of T cells. IncuCyte Zoom (EssenBio) was used to image the wells every 15 min for 40 h at 37 °C and 5% CO2. The data were used to produce time-lapse videos of T-cell cytotoxicity using IncuCyte Zoom software.
FRET peptide cleavage
Peptides containing protease cleavage sequences were synthesized containing an N-terminal Dabcyl and C-terminal EDANS (Genscript). Peptides were dissolved in DMSO at 10 mM. Peptides were further diluted to 100 μM in PBS and mixed with 1 × 105 tumor cells to a final concentration of 10 uM in a total volume of 200 pl in 96-well flat-bottom plates. Plates were incubated overnight at 37 °C in a fluorescence plate reader (Bio-Tek), with fluorescence readings taken every 5 min at excitation and emission wavelengths of 340 and 485 nm, respectively.
Prediction of HLA-peptide interactions
Single HLA allele immunopeptidome data were sourced from Abelin et al.22, who introduced transgenes encoding each allele into MHC-I-deficient cells, immunoaffinity-purified peptide-HLA complexes and determined peptide sequences by mass spectrometry. HLA binding affinities were predicted using MHCflurry (v.1.2.2), predicting against the relevant single-expressed HLA allele. Peptide-MHC stability values were calculated using netMHCstab (v.1.0)32 for each allele where stability prediction was available.
Antibodies
Clinically available antibodies Rituxan (Anti-Human CD20, Roche) and Erbitux (Anti-Human Epidermal Growth Factor Receptor, Merck) were used for conjugation experiments. Antibodies used for flow cytometric analysis were acquired from BioLegend.
Tumor cell dissociation
After excision, tumors were washed with PBS and resuspended in 1X collagenase/hyaluronidase solution (StemCell Technologies) and incubated at 37 °C for 3 h. The dissociated cells were pelleted and resuspended in 0.25% trypsin (Corning) for 5 min before filtering through a 70-μm membrane (BD Biosciences). Cells were washed in PBS and filtered a second time before being labeled with antibodies for flow cytometric analysis.
Mice
NSG mice were purchased from The Jackson Laboratory and used in studies when 6–8 weeks old. C57BL/6, Ccr2−/−, NOD/SCID and OT-I mice were originally obtained from The Jackson Laboratory, and bred and maintained in the Cox-7 gnotobiotic animal facility at Massachusetts General Hospital (MGH). All animal work was conducted under the approval of the MGH Institutional Animal Care and Use Committee and in accordance with federal, state and local guidelines.
In vivo activation and cytotoxicity of T cells
NSG mice (n = 3) received an injection of 5 × 106 tumor cells (MDA-MB-231) orthotopically. After tumors had grown to an appropriate volume, all mice were given 0.5 × 106 freshly isolated CMV peptide-specific T cells intratumorally. After 24 h, the tumors were excised and dissociated (as above). Cells were stained for T-cell activation markers (CD25 and CD38), as well as with CD107a,b for T-cell cytotoxic function, and labeled cells were analyzed by flow cytometry.
In vivo efficacy using primary tumor model
NOD/SCID or NSG mice received an injection of 5 × 106 tumor cells (MDA-MB-231 and SNU-475 cells orthotopically and lung cancer PDX cells (MGH-1088) subcutaneously). After tumors had grown to an appropriate volume, all mice were given 106 ex vivo expanded CMV peptide- specific T cells (as shown in Supplementary Fig. 6) via tail vein injection on a weekly basis for the period shown. Mice were randomly assigned to a group and treated with either 100 pg of APECs containing an irrelevant peptide or different concentrations (50/100 pg per dose intraperitoneally) of APECs containing the relevant peptide and 20,000 IU of recombinant human IL-2 (Peprotech) via intraperitoneal injection. Repeat doses of both APECs and IL-2 were administered three times per week intraperitoneally, and repeat doses of T cells administered once per week (intravenously) for 4 weeks. Tumor size was determined by caliper measurement as a volume three times weekly, and mice were euthanized when tumor volume exceeded 1,200 mm3.
In vivo efficacy using metastatic tumor model
NSG mice received an injection of 5 × 106 MDA-MB-231 cells directly into the mammary fat pad, and tumor cells were allowed to grow until they had reached 8 × 8 mm2 in size. At this time, all mammary fat pad tumors were resected and mice started treatment 3 d post-resection. All mice received 106 ex vivo expanded CMV peptide-specific T cells (as shown in Supplementary Fig. 6) via tail vein injection. Mice were split into two groups, with one group receiving 100-μg intraperitoneal injection of APECs containing an irrelevant peptide and the second group receiving 100- μg intraperitoneal injection of APECs containing the relevant peptide cells and 20,000 IU of recombinant human IL-2 (Peprotech). Repeat doses of both APECs and IL-2 were administered three times per week intraperitoneally, and repeat doses of T cells administered once per week (intravenously) for 4 weeks.
In vivo efficacy using immunocompetent mouse CRC liver metastasis model
To model liver metastasis in the host with intact systemic immunity, the spleen of a C57BL/6 or Ccr2−/− mouse was split in two sections and 1 × 105 SL4 cells were injected into the distal caudal sector, which was then resected30. The other hemi-spleen remained in place. Tumor burden was assessed by measuring Gaussia luciferase (Gluc) activity in the blood from GUc-transduced SL4 tumorbearing mice30. After the development of macroscopic liver metastases (~7 d post-injection), mice were randomly assigned to treatment groups of various combinations of the following treatments as indicated and treatment was initiated: anti-VEGF therapy (B20–4.1.1, 5 mg kg−1, 2 × weekly, generous gift from Genentech); ICB (anti-PD-1 100 μg + CTLA-4 200 μg, 2 × weekly); and anti-murine EGFR D1D4J -MMP14-SIINFEKL-APEC (1.67 mg kg−1, 2 × weekly for 2 weeks), with adoptive transfer of 106 ex vivo expanded OT-I CD8+ T cells at one time. Mice were euthanized at the designated time points for sample collection or when they become moribund. Survival was estimated by Kaplan-Meier curves and compared using the log-rank test.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data presented in this study are available in the article, Supplementary Information or from the corresponding author on reasonable request.
Supplementary Material
Suppl.Video 5:Breast cancer cells labeled with an MMP2-cleavable cetuximab-APECs are killed by peptide-specific CMV-CTLs. An MMP2-cleavable cetuximab APEC was used to label the breast cancer cell line MDA-MB-231. After co-incubation of labeled tumor cells with CMV-CTLs, tumor cell lysis occurred as T cells recognized the CMV epitope being presented at the cell surface by tumor cells.
Suppl.Video 3:Cetuximab-labeled breast cancer cells are not recognized by CMV-CTLs. Breast cancer cells (MDA-MB-231) were labeled with the anti-EGF receptor antibody cetuximab and incubated with CMV-CTLs. There was no recognition of tumor cells by CMV-CTLs.
Suppl.Video 1Untreated breast cancer cells do not instigate CMV-CTL cytotoxicity. Real-time imaging of MDA-MB-231 cell line containing a red nuclear dye to determine cytotoxicity by NLV-specific CMV-CTLs over 40 h. Untreated MDA-MB-231 cells were not targeted by CMV-CTLs during co-culture.
Suppl.Video 2:Breast cancer cells exogenously labeled with CMV peptide are killed by peptide-specific CMV-CTL. Breast cancer cells (MDA-MB-231) were exogenously labeled with CMV peptide (NLVPMVATV) and co-incubated with peptide-specific CMV-CTLs. During co-culture, CMV-CTLs recognized the peptide displayed by the tumor cells and efficiently killed the breast cancer cells.
Suppl.Video 4:Negative control cetuximab-APEC-labeled breast cancer cells are not recognized by CMV-CTLs. A negative control cetuximab APEC, designed to prevent release of the CMV epitope, was used to label the breast cancer cell line MDA-MB-231. After co-incubation of labeled tumor cells with CMV-CTLs, tumor cells were not targeted for killing by T cells.
Acknowledgements
D.G.D’s work was supported through NIH grant no. R41-CA213678, the Proton Beam/Federal Share Program and MGH ECOR. We thank P Huang, S. Roberge and T. Hbatmu for the maintenance of gnotobiotic animal colonies and experimental assistance. This work was supported by NCI program project grant nos. P01-CA080124 and R01-CA208205 (to R.K.J. and D.F.), NCI grant nos. R35-CA197743 and U01-CA 224348 (to R.K.J.) and also in part by the Ludwig Center at Harvard. The NIH Tetramer Facility is supported by contract HHSN272201300006C from the National Institute of Allergy and Infectious Diseases, a component of the National Institutes of Health in the Department of Health and Human Services.
Competing interests
R.K.J. received an honorarium from Amgen and consultant fees from Ophthotech, SPARC, SynDevRx and XTuit. R.K.J. owns equity in Enlight, SPARC and SynDevRx, and serves on the Board of Directors of XTuit and Boards of Trustees of Tekla Healthcare Investors, Tekla Life Sciences Investors, Tekla Healthcare Opportunities Fund and Tekla World Healthcare Fund. D.G.D. received consultant fees from Bayer and BMS and has research grants from Bayer, Merrimack, Leap, Exelixis and BMS. M.C. and D.G.M. are named inventors on patent application no. WO 2012/123755, which is licensed to Revitope Oncology. D.G.M. and M.C. own equity in Revitope Oncology. M.C. holds equity in Gritstone Oncology. M.C. received consultant fees from Merck Laboratories. M. C. is now an employee of AstraZeneca.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41587-019-0404-8.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41587-019-0404-8.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brahmer JR et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med 366, 2455–2465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yarchoan M, Hopkins A & Jaffee EM Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med 377, 2500–2501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le DT et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown SD et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 24, 743–750 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Z, Ott PA & Wu CJ Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat. Rev. Immunol 18, 168–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence MS et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellmann MD et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med 33, 853–861.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fridman WH, Pages F, Sautes-Fridman C & Galon J The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer 12, 298–306 (2012). [DOI] [PubMed] [Google Scholar]
- 9.McGranahan N et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentzen AK et al. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat. Biotechnol 34, 1037–1045 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Carreno BM et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahin U et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Ott PA et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan RA et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 314, 126–129 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grupp SA et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med 368, 1509–1518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.June CH & Sadelain M Chimeric antigen receptor therapy. N. Engl. J. Med 379, 64–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karrer U et al. Memory inflation: continuous accumulation of antiviral CD8+ T cells over time. J. Immunol 170, 2022–2029 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Khan N et al. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J. Immunol 169, 1984–1992 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Reddehase MJ Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol 2, 831–844 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Simoni Y et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Scheper W et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med 25, 89–94 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Abelin JG et al. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perosa F et al. Beta 2-microglobulin-free HLA class I heavy chain epitope mimicry by monoclonal antibody HC-10-specific peptide. J. Immunol 171, 1918–1926 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Edwards DR The Cancer Degradome: Proteases and Cancer Biology (Springer, 2008). [Google Scholar]
- 25.Desnoyers LR et al. Tumor-specific activation of an EGFR-targeting probody enhances therapeutic index. Sci. Transl. Med 5, 207ra144 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Kantarjian H et al. Blinatumomab versus chemotherapy for advanced acute lymphoblastic leukemia. N. Engl. J. Med. 376, 836–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Rourke DM et al. A single dose of peripherally infused EGFRvIII- directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med 9, pii: eaaa0984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka A & Sakaguchi S Regulatory T cells in cancer immunotherapy. Cell Res. 27, 109–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nangia V et al. Exploiting MCL1 dependency with combination MEK + MCL1 Inhibitors leads to induction of apoptosis and tumor regression in KRAS-mutant non-small cell lung cancer. Cancer Discov. 8, 1598–1613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahbari NN et al. Anti-VEGF therapy induces ECM remodeling and mechanical barriers to therapy in colorectal cancer liver metastases. Sci. Transl. Med 8, 360ra135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosato PC et al. Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat. Commun 10, 567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Donnell TJ et al. MHCflurry: open-source class I MHC binding affinity prediction. Cell Syst. 7, 129–132 e124 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl.Video 5:Breast cancer cells labeled with an MMP2-cleavable cetuximab-APECs are killed by peptide-specific CMV-CTLs. An MMP2-cleavable cetuximab APEC was used to label the breast cancer cell line MDA-MB-231. After co-incubation of labeled tumor cells with CMV-CTLs, tumor cell lysis occurred as T cells recognized the CMV epitope being presented at the cell surface by tumor cells.
Suppl.Video 3:Cetuximab-labeled breast cancer cells are not recognized by CMV-CTLs. Breast cancer cells (MDA-MB-231) were labeled with the anti-EGF receptor antibody cetuximab and incubated with CMV-CTLs. There was no recognition of tumor cells by CMV-CTLs.
Suppl.Video 1Untreated breast cancer cells do not instigate CMV-CTL cytotoxicity. Real-time imaging of MDA-MB-231 cell line containing a red nuclear dye to determine cytotoxicity by NLV-specific CMV-CTLs over 40 h. Untreated MDA-MB-231 cells were not targeted by CMV-CTLs during co-culture.
Suppl.Video 2:Breast cancer cells exogenously labeled with CMV peptide are killed by peptide-specific CMV-CTL. Breast cancer cells (MDA-MB-231) were exogenously labeled with CMV peptide (NLVPMVATV) and co-incubated with peptide-specific CMV-CTLs. During co-culture, CMV-CTLs recognized the peptide displayed by the tumor cells and efficiently killed the breast cancer cells.
Suppl.Video 4:Negative control cetuximab-APEC-labeled breast cancer cells are not recognized by CMV-CTLs. A negative control cetuximab APEC, designed to prevent release of the CMV epitope, was used to label the breast cancer cell line MDA-MB-231. After co-incubation of labeled tumor cells with CMV-CTLs, tumor cells were not targeted for killing by T cells.
Data Availability Statement
Data presented in this study are available in the article, Supplementary Information or from the corresponding author on reasonable request.