Figure 3.
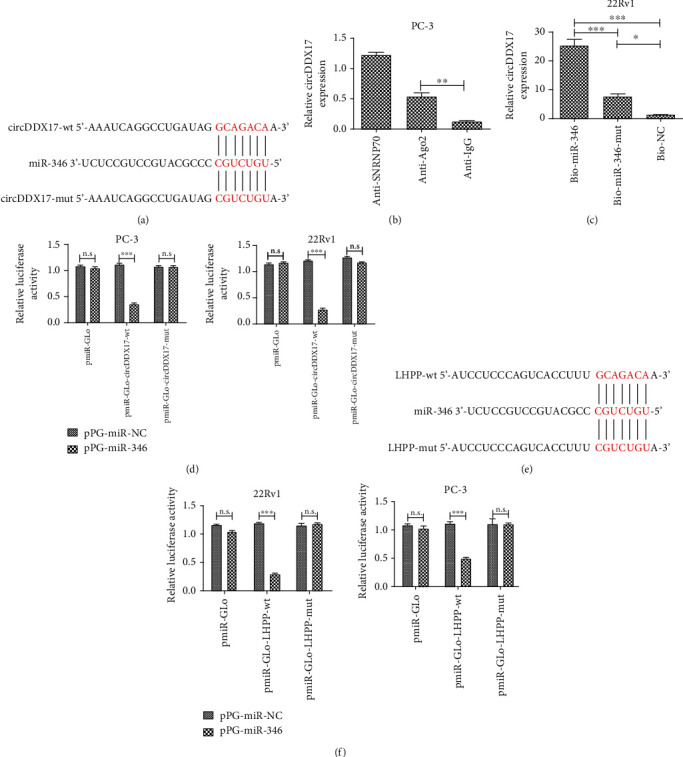
Identification of potential matching sequence in miR-346 and LHPP mRNA. (a) Alignment of miR-346 sequence with circDDX17 and circDDX17 mutated at the potential binding site. (b) Relative luciferase activity in 22Rv1 and PC-3 cell lines cotransfected with pPG-miR-138 (or the empty vector as a control) and the luciferase empty vector (pmiR-GLo) or the vector containing the wild-type circDDX17 (pmiR-GLo-DDX17-wt) or mutant transcripts (pmiR-GLo-DDX17-mut) was detected. All data are shown as the relative ratio of firefly luciferase activity to Renilla luciferase activity. (c) PC-3 cell lines were transfected with biotinylated NC (Bio-NC), biotinylated wild-type miR-346 (BiomiR-346), or biotinylated mutant miR-346 (Bio-miR-346-mut), and biotin-based miRNA pull-down assays were performed after 48 h of transfection. circDDX17 levels were determined by qPCR. (d) Amount of circDDX17 bound to SNRNP70 (a positive control), Ago2, or IgG (a negative control) was detected by qPCR after RIP in 22Rv1 cells. (e) Alignment of LHPP mRNA sequence with miR-346 and miR-346 mutated at the potential binding site. (f) Relative luciferase activity in 22Rv1 and PC-3 cell lines cotransfected with pPG-miR-346 (or the empty vector as a control) and the luciferase empty vector (pmiR-GLo) or the vector containing the wild-type LHPP (pmiR-GLo-LHPP-wt) or mutant transcripts (pmiR-GLo-LHPP-mut) was detected. All data are shown as the relative ratio of firefly luciferase activity to Renilla luciferase activity. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
