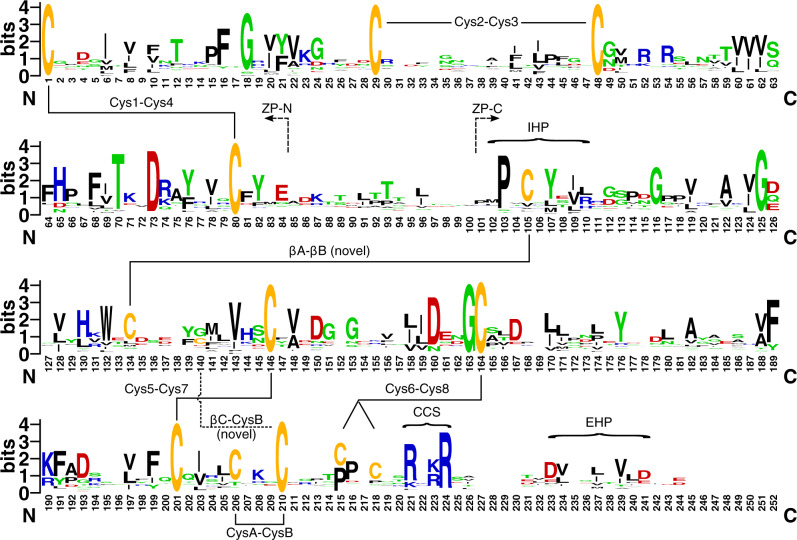
Fig. 1.
Nematode ZP module amino acid conservation patterns. Residue height indicates its prevalence in the top-scoring alignment. Connections between cysteine residues indicate inferred disulfide linkages; also shown are the approximate boundaries of the ZP-N and ZP-C domains, the internal and external hydrophobic patches (IHP/EHP), and the consensus cleavage site (CCS). Nonhomologous flanks and insertions were trimmed from the sequences as part of the alignment process; the relationship between alignment numbering and untrimmed sequence position for untrimmed Caenorhabditis elegans CUT-1 is provided in supplementary figure 1, Supplementary Material online.