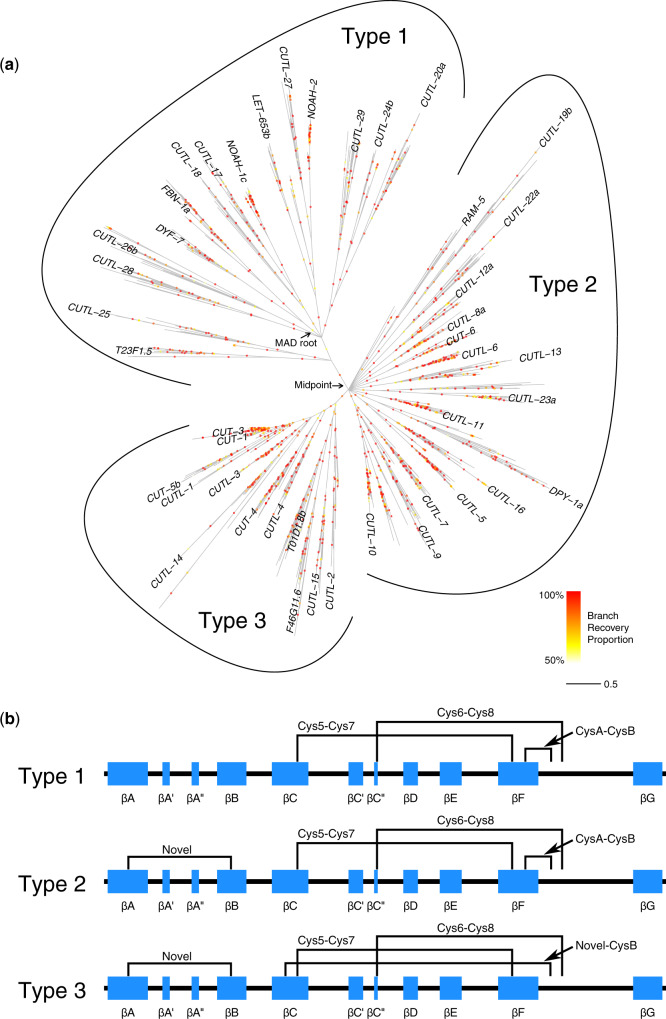
Fig. 2.
Nematode ZP module phylogeny. (a) The majority-rule consensus of ML phylogenies estimated for 100 replicate ZP module alignments. BRPs are shown using colored circles; darker/redder circles indicate greater robustness to alignment variation. Branch lengths, drawn in amino acid substitutions per site (see scale bar), were estimated via ML using the top-scoring alignment. The labeled arrows indicate the Minimal Ancestor Deviation (MAD) root position and the phylogenetic midpoint. Tip names are shown for Caenorhabditis elegans ZP modules; for clarity, CUT-1 was moved slightly to avoid overlap with CUT-3. Three major subtrees are noted (Type 1/2/3), the members of which are defined by different ZP-C domain cysteine connectivity patterns. (b) Cysteine connectivity patterns for Type 1/2/3 ZP-C domains, inferred based on amino acid conservation patterns and homology modeling of C. elegans ZPD proteins. The β-strand secondary structure diagram follows that of the human uromodulin ZP-C domain (Bokhove et al. 2016). The position of the MAD root in (a) suggests that the Type 1 connectivity pattern represents the ancestral state.