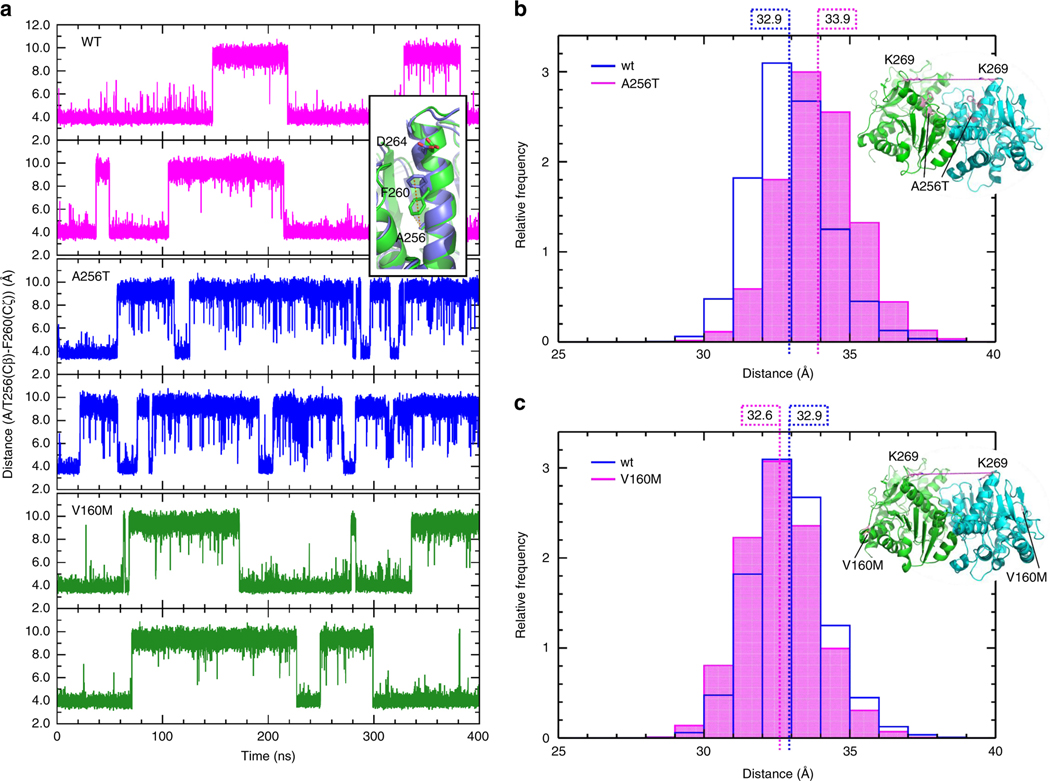
Fig. 3. Molecular dynamics simulations in MDH2 p.Ala256Thr and p.Val160Met variants of unknown significance (VUS) versus wild-type (WT).
a Distance between the Cζ atom of F260 and the Cβ atom of A/T256 for both monomers of WT MDH2 (magenta, upper panels), Ala256Thr (blue, middle panels), and V160M (green, lower panels) during the simulations. A short distance corresponds to the F260 down conformation, and a long distance to the F260 up conformation (inset). b and c Distribution of the distance between the Cα atoms of K269 of the two monomers of the a Ala256Thr and b Val160Met variants from snapshots from the simulation trajectories, compared with WT