Figure 2: PAX–FOXO1 fusion gene drives RMS formation.
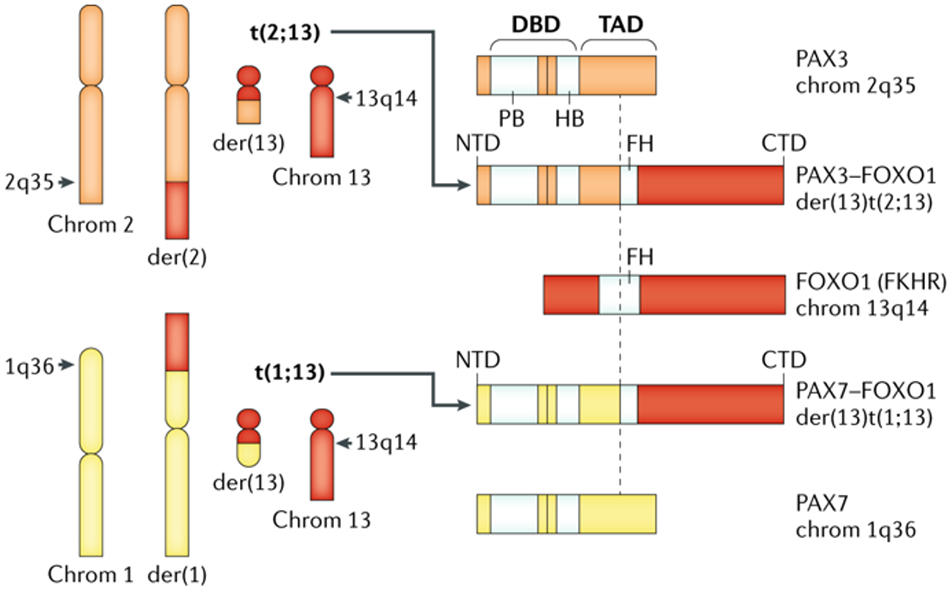
FP RMS is defined by balanced translocations between PAX3, residing in the Giemsa band 35 on the long (q) arm of chromosome 2 (2q35) or PAX7 on chromosome 1 (1q36) with the FOXO1 gene on chromosome 13 (13q14) generating fusion proteins. These balanced translocations generate two derivative (der) chromosomes (left side panel), only one of which encodes for the PAX3 (or PAX7)-FOXO1 fusion mRNA and protein (right side of panel). These fusion proteins contain the amino terminal portion of either PAX3 or, less commonly, PAX7 and the carboxyl portion of FOXO1. The amino terminus of the fusion protein includes motifs needed for DNA binding from the respective PAX gene, and the carboxyl terminus of the fusion protein is felt to alter the transcriptional activation domain of the oncogenic transcription factor. Note that the alternate derivative chromosomes are not known to contribute to RMS pathogenesis. Chrom, chromosome; FP, fusion positive; RMS rhabdomyosarcoma; der; derivative chromosome; DBD, DNA binding domain; TAD, transcriptional activation domain; PB, paired-box domain; HB, homeobox domain; FH, Forkhead-related domain; FKHR, forkhead homolog in RMS (original designation of FOXO1 gene).
