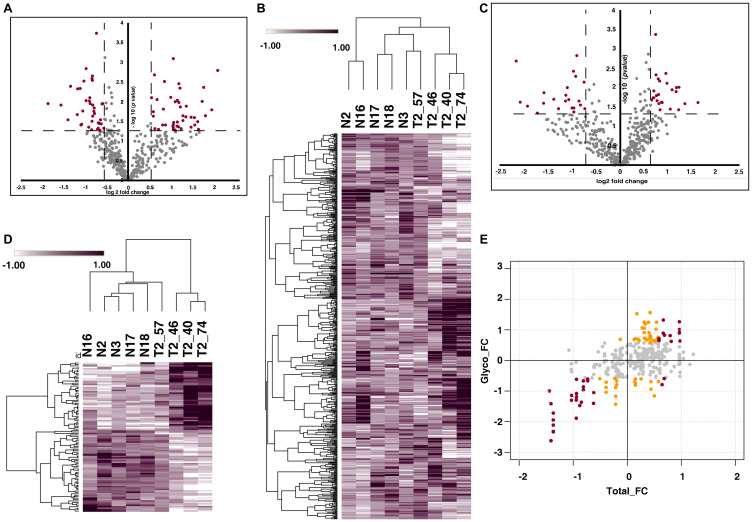
Figure 3. Urinary N-glycoproteome and proteome of MIBC.
(A) Volcano plot shows the N-glycoproteome profile of MIBC and healthy subjects. Fold change of the N-glycosylation abundances (log2) are plotted against the t-test p-values (-log10). Significance levels are indicated by dashed lines. (B) Heat map representing the significantly glycosylated peptides in MIBC group in comparison with the healthy subjects (p-value < 0.05). Abundance values of the differentially glycosylated peptides are represented in the heat map. (C) Volcano plot represents comparative proteomic profile of MIBC and healthy subjects. Fold change of the protein abundances versus t-test p-values (-log10). Significance levels are indicated by dashed lines. (D) Heat map of differentially expressed proteins (log2 abundance value) of MIBC compared with the healthy subjects. (E) Global presentation of proteomic and respective glycosylation occupancy. Yellow datapoints represent differential glycosylation occupancy with fold change ≥ 1.5 and red datapoints with fold change ≥ 2 on proteins that are unchanged in proteomic data. Grey datapoints represent proteins that are unchanged in both datasets.