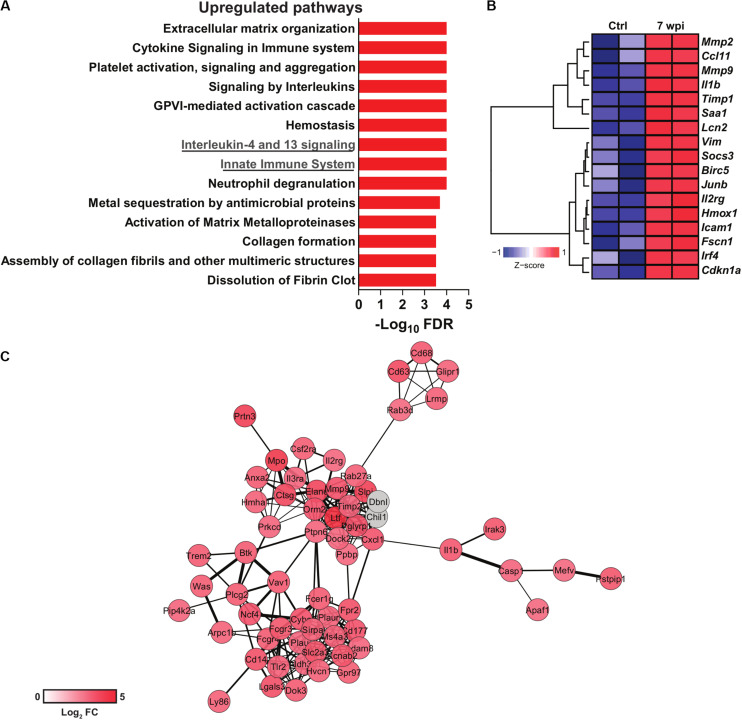
FIGURE 1.
Transcriptomics data reuse of mice infected with S. japonicum. Normalized gene expression data from livers of mice infected with S. japonicum for 7 weeks and naïve mice were acquired at the Gene Expression Omnibus (GEO) repository (GSE14367). Differential gene expression was evaluated with the package limma for R. (A) Pathway enrichment analysis of upregulated genes in liver of S. japonicum-infected mice. Pathways were evaluated with the ToppGene suite platform (Chen et al., 2009) and the bar plot depicts the significance of enrichment in each pathway as shown by -log10 p-values adjusted by Benjamini-Hochberg false discovery rate (FDR) method. (B) Heat map showing the expression of genes included in the interleukin-4 and 13 signaling pathway in S. japonicum-infected and naïve mice. Hierarchical clustering was performed with the package amap for R using Euclidian distance metric and Ward linkage. The heat map was generated with the package gplots for R. (C) Gene network composed of genes included in the innate immune system pathway. The network was generated with the platform STRING v11.0 (Szklarczyk et al., 2015) and visualized with Cytoscape v3.7.2 (Shannon et al., 2003). The color scale is proportional to the log2 fold changes of each gene from S. japonicum-infected mice over naïve mice. Genes colored in gray were imputed by STRING but were not present in the evaluated dataset.