Abstract
Neuronal polarization and growth are developmental processes that occur during neuronal cell differentiation. The molecular signaling mechanisms involved in these events in in vivo mammalian brain remain unclear. Also, cellular events of the neuronal polarization process within a given neuron are thought to be constituted of many independent intracellular signal transduction pathways (the “tug-of-war” model). However, in vivo results suggest that such pathways should be cooperative with one another among a given group of neurons in a region of the brain. Lipid rafts, specific membrane domains with low fluidity, are candidates for the hotspots of such intracellular signaling. Among the signals reported to be involved in polarization, a number are thought to be present or translocated to the lipid rafts in response to extracellular signals. As part of our analysis, we discuss how such novel molecular mechanisms are combined for effective regulation of neuronal polarization and growth, focusing on the significance of the lipid rafts, including results based on recently introduced methods.
Keywords: growth cone, lipid rafts, phosphoproteomics, JNK, super-resolution microscopy, palmitoylation
Introduction
Brain development in mammals is believed to involve six steps, including: (1) segmentation of brain regions; (2) neuronal differentiation from neural stem cells; (3) neuronal migration to the appropriate locations; (4) neuronal polarity determination and axon growth as directed by guidance molecules; (5) synaptogenesis; and (6) removal of excess synapses (Sanes et al., 2019). Except for the last step, which depends on neuronal activity, the other steps appear to be regulated by genetic mechanisms. In this review article, we focus on molecular aspects of the fourth step of the above sequence of mammalian brain development.
More than 30 proteins have been characterized based on their involvement in neuronal polarization at the single-cell level (Takano et al., 2019). While many of these proteins likely contribute to neuronal polarization in similar ways, these molecules were discovered in independent studies, and little is known about how these proteins might act in a coordinated fashion. In this review, we focus on the potential role of lipid rafts in neuronal polarization and axon growth (Igarashi, 2019).
Lipid Rafts: What Is Important for Signaling?
Glycerophospholipids are major components of the plasma membrane, and membrane proteins are incorporated in such lipids (Brown and London, 1998; Lorent and Levental, 2015). According to the classical model, such lipids have high fluidity, behaving like a liquid, due to the unsaturated fatty acids bound to these phospholipids; all of the membrane proteins thus would be flowing in a “sea” of membrane lipids, freely diffusing to anywhere within the membrane. In contrast to such an idea (that the membrane structure is uniform), the concept of the lipid rafts has been postulated. Namely, minor components of the membrane lipids, including cholesterol (sterol) and sphingolipids (sphingomyelin and glycolipids such as gangliosides), are present in a concentrated and clustered form in specific domains of the membrane. Biophysical properties of these minor membrane lipids predict that the lipid raft domain has much lower fluidity than that of the major components (the glycerophospholipids; Lorent and Levental, 2015). These low fluidity regions serve as anchors for specific membrane proteins that reside therein, and the lipid rafts are thought to be “signaling hotspots” for responding to extracellular signals (Lingwood and Simons, 2010; Egawa et al., 2018).
Two types of membrane proteins are thought to be specifically associated with lipid rafts: glycosylated phosphatidylinositol (GPI)-anchored proteins (Saha et al., 2016) and palmitoylated proteins. GPI-anchored proteins are located at the cell surface and are attached to the plasma membrane with a GPI anchor, but cannot directly interact with intracellular signaling proteins; thus, GPI-anchored proteins require co-receptors that possess transmembrane domains. GPI-anchoring sugar chains are synthesized in the endoplasmic reticulum (ER) and then undergo fatty acid modification at PI within the Golgi apparatus before being sorted to the plasma membrane (Saha et al., 2016). The resulting GPI-anchored proteins have been observed to repeatedly undergo rapid gathering and scattering within lipid rafts (Suzuki et al., 2017; Figure 1).
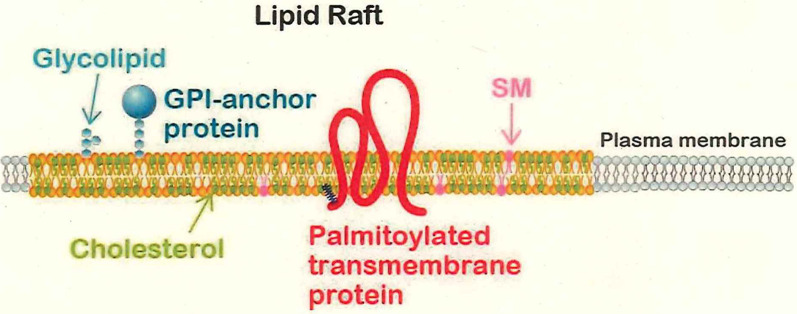
Figure 1.
The lipid raft domain. Lipid rafts are composed of sphingolipids such as glycolipids and sphingomyelin (SM), cholesterol, and glycosylated phospholipid (GPI)-anchored or palmitoylated membrane proteins. Lipid rafts are thought to be interspersed among non-raft domains that are composed of the glycerophospholipids and exhibit high fluidity. The lower fluidity of the lipid rafts is presumed to lead to retention and localized concentration of membrane proteins that participate in signal transduction in response to extracellular signals.
Protein palmitoylation is an S-acylation modification of clustered cysteine residues; this protein modification is performed in the Golgi apparatus (Chini and Parenti, 2009; Resh, 2016). For soluble proteins, palmitoylation simply endows the targets with an affinity for the plasma membrane; for transmembrane proteins, palmitoylation is believed to direct the targets for sorting to the lipid raft domains (Stepanek et al., 2014; Lorent and Levental, 2015). More than 20 palmitoyltransferases have been identified in mammals, and each of these enzymes is thought to have specific physiological substrates (Fukata et al., 2016). Of the major palmitoylated proteins in the adult brain, more than half are transmembrane proteins (Kang et al., 2008). Previous work has demonstrated the significance of palmitoylated transmembrane proteins in neurobiology (Vallejo et al., 2017; Hayashi, 2020); the relationship of palmitoylation to neuronal polarity will be addressed later in this review article.
Neuronal Polarization and the Need for Its Rapid Determination
The selection of the specific regions of a neuron where growth cones form is an important problem; neuronal polarity is key to the formation of the axon (a single output process) and dendrites (multiple input processes; Laumonnerie and Solecki, 2018). Neuronal polarity determination has been classified into five stages (Dotti et al., 1988), namely, stage 1: initiation of the emergence of the minor process(es); stage 2: the growth of the minor processes; stage 3: axon specification; stage 4: dendritic specification; and stage 5: synaptogenesis. Among these steps, the transition from stage 2 to stage 3 has been the most intensively studied (Funahashi et al., 2020). Most of the previous studies on the establishment of neuronal polarity have examined cell-autonomous signaling pathways in individual (single) cells in in vitro culture systems (Funahashi et al., 2020). Based on these previous studies, a tug-of-war model (Lalli, 2014) has been adopted to explain neuronal polarization. This model (Figure 1) is based on the experimental facts that although each neuron in dissociation culture (particularly when grown on artificial culture substrates) has an intrinsic mechanism for neuronal polarization, at stage 2, each minor neuronal process performs the inter-dependent interactions for signaling in a tug-of-war. After spending a relatively long time (~48 h) at stage 2, the model explains that the sole process that “wins” this “tug of war” requires rapid growth to differentiate successfully into an axon at stage 3 (Lalli, 2014; Guo and Cheng, 2015).
However, it seems unlikely that the signaling leading to polarization of neurons occurs spontaneously under in vivo conditions (Namba et al., 2014). For in vivo mammalian brain development, each neuron within a group would have to acquire polarity simultaneously, and then also grow an axon simultaneously in the same direction, a series of events that seems far more complicated than the simple tug-of-war mechanism. Namely, in vivo, stage 2 (a stage of undecided polarity) cannot persist for an extended interval, and the transition from stage 2 to stage 3 (a stage of defined polarity) cannot proceed in a disorderly fashion. It is difficult to imagine that intrinsic factors alone would be expressed in vivo in a large number of the neurons just before stage 3 in a manner that would permit (despite the restricted time course) synchronization of the polarization with the axon growth direction (Namba et al., 2014). The mechanisms of stage 3 itself (rapid axon growth) sometimes appear to conflict with those proposed for the transition from stage 2 to stage 3 (Takano et al., 2019; Figure 2A).
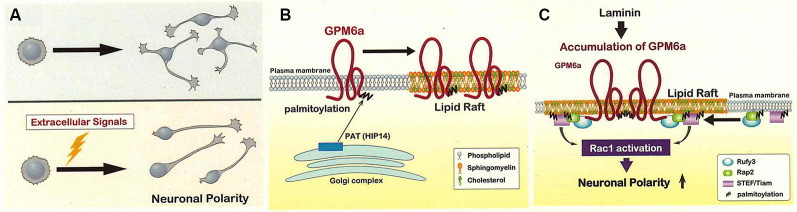
Figure 2.
Lipid rafts may be the site of signaling for the determination of the neuronal polarity. (A) The polarity of each neuron is probably not determined in an inconsistent way (upper) but instead is synchronized in vivo by extracellular signals exchanged among neurons (lower). The lower mechanism is expected to shorten the time for polarity determination compared to the upper one. (B) Glycoprotein M6a (GPM6a) is palmitoylated and sorted to the lipid rafts in the neuronal plasma membrane. GPM6a is believed to be palmitoylated at cysteine clusters (located near the protein’s N-terminus) via a reaction catalyzed by the HIP14 or ZHHC17 palmitoyltransferases; modification would occur within the Golgi apparatus, and GPM6a then would be inserted into the lipid raft domains (Butland et al., 2014). Although the non-palmitoylated form of GPM6a is localized to non-raft domains of the plasma membrane, this form of GPM6a does not appear to mediate biological effects in response to extracellular signals (Honda et al., 2017a). (C) Laminin induces the assembly of signaling molecules downstream of GPM6a around lipid rafts, an event that contributes to the rapid determination of polarity (see Honda et al., 2017a). GPM6a, Rap2, and Tiam2 are present in the lipid rafts with Rufy3, an adaptor protein that acts as a linker between GPM6a and Rap2-Tiam2 (Honda et al., 2017a,b). Tiam2 is a guanine nucleotide exchange factor (GEF) that activates Rac and is expected to contribute to the rapid determination of polarity. Modified from Honda et al. (2017a).
Thus, there appears to be a role for extrinsic factors in inducing polarity determination within the neuronal population. Candidates for such signals have been identified (Takano et al., 2019), and include extracellular matrix components such as laminin (LN), a protein that is highly abundant in the developing brain (Esch et al., 1999; Randlett et al., 2011; Johnson et al., 2012; Honda et al., 2017a,b; Serjanov et al., 2018). LN facilitates neuronal polarity determination, as demonstrated by the ability of exogenously supplied LN to permit neurons to “skip” stage 2 of development (Honda et al., 2017a,b).
Neuronal Polarization Related to Lipid Rafts
Signaling Molecules for Polarization in Lipid Rafts
Among the many proteins involved in neuronal polarization (Takano et al., 2019), more than 10 species that are present upstream of the signaling have been reported to be present in lipid rafts or to be translocated to lipid rafts when the corresponding signals are activated (Table 1). These results suggest that those molecules are likely to function in polarization signaling as the concentrated forms in lipid rafts.
Table 1.
Proteins reported to localize to lipid rafts or to be translocated to lipid rats in response to extracellular stimuli.
| A. Receptors and cell adhesion molecules |
| TrkB (Assaife-Lopes et al., 2010; Mandyam et al., 2017) |
| IGF-1R (Sural-Fehr et al., 2019) |
| Neuropilin/Plexin complex (Dang et al., 2012) |
| Integrin (Decker et al., 2004) |
| Thy-1 (Ledesma et al., 1998) |
| B. Protein kinases |
| CaMKI (Davare et al., 2009) |
| Glycogen synthase kinase-3 (Sui et al., 2006) |
| SAD-B (Rodríguez-Asiain et al., 2011) |
| Akt (Bryant et al., 2009) |
| Fyn (Ko et al., 2005) |
| C. Other intracellular signaling molecules |
| PI3K (Zheng et al., 2014) |
| Wnt-Dvl (Frizzled; Haack et al., 2015) |
| Ras/Rap (Zhang et al., 2018) |
| Rac1 (Fujitani et al., 2005; Grider et al., 2009; Köster et al., 2014; Lee et al., 2016) |
| V-ATPase (Kanda et al., 2013; Makdissy et al., 2018) |
The proteins involved in polarization are listed above.
Neuronal polarization is known to depend on the positioning of the Golgi apparatus, and thus, the biochemical mechanisms in that organelle should have important effects on this event (Villarroel-Campos et al., 2016; Tortosa and Hoogenraad, 2018; Caracci et al., 2019). Although such biochemical processes are not completely understood, one essential modification performed in the Golgi apparatus is protein palmitoylation, which regulates the trafficking of proteins for axon specification (Rodríguez-Asiain et al., 2011; Tortosa et al., 2017; Tortosa and Hoogenraad, 2018). In mammals, protein palmitoyltransferases (PATs, the enzymes responsible for this reaction) are concentrated in the cis-Golgi and catalyze S-palmitoyl acylation of cysteine residues in target proteins (Ernst et al., 2019). Such fatty acylation of soluble proteins is believed to recruit these proteins to the plasma membrane. This modification also is employed for membrane proteins, although the purpose of palmitoylation of such proteins (which are already membrane-associated) is less apparent. Notably, however, in various cells (including the neuron), palmitoylation increases recruitment of such transmembrane proteins to lipid rafts (Linder and Deschenes, 2007; Hayashi, 2020). Palmitoylation is thought to modify the membrane trafficking of the target proteins, possibly by changing the curvature of the sorting vesicles carrying these proteins (Ernst et al., 2019).
In the adult rodent brain, more than 20 species of major palmitoylated proteins have been identified; more than half are transmembrane proteins, a class that includes Glycoprotein M6a (GPM6a; Kang et al., 2008).
GPM6a Signaling in Response to LN
GPM6a, a potential regulator of neuronal growth, is a major membrane protein of the growth cone (Nozumi et al., 2009); specifically, GPM6a is a four-transmembrane-domain protein that is known to be highly expressed in differentiated neurons (Möbius et al., 2008). This gene product is a major palmitoylated protein in the adult brain (Kang et al., 2008). Although GPM6a’s exact roles remained unclear, we suspected that this protein might be a signal transducer for LN-dependent signaling. Notably, inhibition of GPM6a palmitoylation abolished LN-dependent determination, indicating that the trafficking of this protein to lipid rafts is essential to GPM6a’s mechanism of action (Honda et al., 2017a,b), even though GPM6a, being an intrinsic membrane protein already localizes to the plasma membrane (Ito et al., 2018).
Using proteomics, a GPM6a-Rufy3-Rap2a-Tiam2 complex was identified in lipid rafts (Honda et al., 2017a). Rufy3 (also called Singar 1; Mori et al., 2007) and Tiam2/STEF both are known to be involved in neuronal polarization. Tiam2, a Rac guanine nucleotide exchange factor (GEF), determines the site of axon extension via the rapid accumulation of the GTP-bound form of Rac1 (Nishimura et al., 2005). This accumulation of GTP-Rac1 may be useful for organizing multiple otherwise-unrelated signaling molecules that contribute to polarization. For example, the activation of Rac1 by positive feedback in vivo is probably essential to speedy polarization (Acevedo and González-Billault, 2018; Dupraz et al., 2019; Takano et al., 2019); proximity to members of the Tiam family (proteins that serve as Rac GEFs) would facilitate this process. It is physiologically conceivable that Rap2 (Bruurs and Bos, 2014), a member of the Ras GTPase family that is highly palmitoylated (Uechi et al., 2009; Baumgart et al., 2010), is present in lipid rafts, such that the presence of activated Tiam2 in the lipid rafts contributes to polarization (Honda et al., 2017a,b).
Rufy3 is (in in vitro experiments) a multiple adapter protein for small GTPases (Fukuda et al., 2011) and has been shown to bind to activated Rap2 (Kukimoto-Niino et al., 2006; Honda et al., 2017a,b). Rufy3 also is involved in neuronal polarity (Mori et al., 2007), for which the only identified related signaling molecule was PI-3-kinase (PI3K; Mori et al., 2007). In in vivo signaling, lipid rafts may connect GPM6a to Rap2-Tiam via Rufy3; indeed, GPM6a can induce the translocation of Rufy3 to lipid rafts (Honda et al., 2017a,b).
Human Neuropsychiatric Diseases and Polarization
GPM6a is known to be a good endogenous substrate of HIP14/Zdhhc17, a palmitoyl acetyltransferase (protein palmitoyl acyltransferase; PAT) implicated in Huntington disease, a human hereditary neurodegenerative disease (Butland et al., 2014; Figure 2B).
Also, GPM6a, Rufy3, Rap2, and Tiam2 (Figure 2C) all have been implicated in studies of important psychiatric diseases, including analyses of human patient neuropathologies and murine models (Funk et al., 2012; Bhattacherjee et al., 2017; Ma et al., 2018; Aberg et al., 2020). Notably, genome-wide association study (GWAS) identified the genes encoding GPM6a and Rufy3 as loci associated with an elevated risk of human schizophrenia and depression, respectively (Ma et al., 2018; Aberg et al., 2020). Since these diseases are thought to be partly due to the genetic lability of some genes in brain development, such results suggest that GPM6a and downstream molecules have physiological roles in the development of neurons and that GPM6a in lipid rafts may be involved in a key step of neuronal morphogenesis.
Membrane Recycling Mechanisms in Lipid Rafts; Newly Observed Using Super-Resolution Microscopy
Technical Merits and Power of Super-Resolution Microscopy for Analysis of Membrane Trafficking
To better understand the role of membrane trafficking in axonal growth, the precise relationship between both cytoskeletal and membrane components must be clarified. Live imaging has greatly contributed to the understanding of such mechanisms (Igarashi et al., 1996; Tamada and Igarashi, 2017; Dubey et al., 2018; Meka et al., 2019). However, live imaging of growing axons has remained a challenge: the vesicles and cytoskeleton in the growth cone are highly crowded, meaning that each labeled structure overlaps with others, impeding discrimination among the various components. Additionally, for conventional confocal microscopy, the diffraction limit of optical microscopy (~200 nm) has precluded precise analyses of vesicles and cytoskeletal structures in growth cones (Igarashi et al., 2018; Schermelleh et al., 2019).
Recently, several types of super-resolution microscopy have been developed (Hauser et al., 2017; Igarashi et al., 2018; Schermelleh et al., 2019). These methods employ fluorescence microscopy devices to observe intracellular molecules, permitting researchers to overcome the optical diffraction limit and achieve resolutions of 50–100 nm. These new techniques not only make it possible to observe smaller objects but also facilitate the analysis of densely distributed materials such as vesicles and cytoskeletal components in the growth cone (Nozumi and Igarashi, 2018). Also, super-resolution microscopy provides three-dimensional images and so is superior to confocal microscopy in this context (Igarashi et al., 2018).
One super-resolution technique, structured illumination microscopy (SIM), can visualize the fine structure of cells by calculating the interference (moiré) patterns induced by irradiation with striped-pattern excitation light (Gustafsson, 2008). Using SIM, lateral and axial dimensions of approximately 100 and 300 nm (respectively) can be visualized, making super-resolution microscopy useful for tracking molecular dynamics and movements in live-cell imaging (Demmerle et al., 2017; Richter et al., 2018).
Membrane Recycling in Lipid Rafts Contributes to Axon Growth
Although biochemical evidence for the existence of the lipid rafts accumulated until 2010, the idea of the lipid rafts remained a hypothesis. This challenge remained because the visualization of the lipid raft domains remained impossible up to that time. However, the development of super-resolution microscopy permitted the observed lipid rafts in various cell types, leading to the wider acceptance of this concept (Owen et al., 2012). There are several styles of super-resolution microscopy that use distinct probes; each of these methods has successfully permitted the visualization of lipid rafts (Tobin et al., 2014; Chen et al., 2015; Hartley et al., 2015; Stahley et al., 2016; Gao et al., 2017; Schlegel et al., 2019; Angelopoulou et al., 2020). These new results have contributed to models suggesting possible roles for lipid rafts in multiple cellular pathways (Raghunathan and Kenworthy, 2018).
In the neuron, however, such an approach had not been applied, given the elevated density of cholesterol and sphingolipids, particularly gangliosides, in neural membranes. Only recently has the development of 3D-SIM-type super-resolution microscopy permitted imaging of the dynamic endocytotic processes of the lipid raft domains in the growth cone (Nozumi et al., 2017).
3D-SIM depends on the use of D4 (a molecule derived from bacterial theta toxin that shows specific binding to membrane cholesterol); a fusion of green fluorescent protein (GFP) to D4 (GFP-D4) can be used as a probe for labeling cholesterol (Ohno-Iwashita et al., 2004; Ishitsuka et al., 2011). By combining this probe and super-resolution microscopy, we succeeded in visualizing neuronal membrane lipid rafts (Nozumi et al., 2017). These lipid rafts showed movements similar to those seen for GPM6a itself and clathrin-independent endocytosis at the leading edge. Thus, we infer that the lipid rafts are associated with F-actin bundling at the leading edge, where these structures undergo highly dynamic movements as part of axonal growth (Figure 3).
Figure 3.
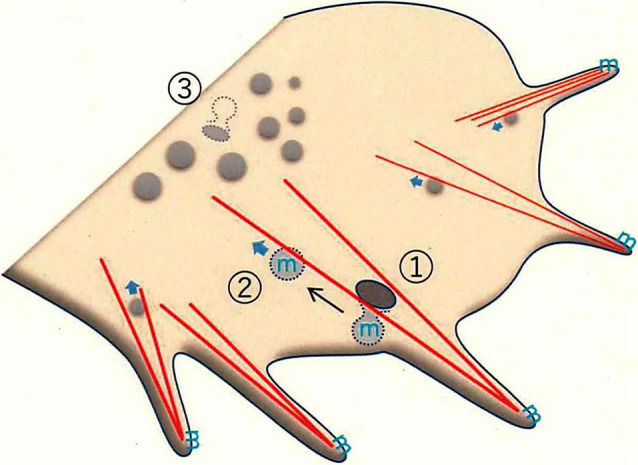
Membrane trafficking in the growth cone, as revealed by super-resolution microscopy. F-actin-dependent endocytosis occurs in the peripheral (P-) domain of the growth cone. The leading edge protrudes as filopodia, which have dense F-actin bundles (red lines). F-actin-bundling for filopodial formation induces endorphin-mediated endocytosis (EME; ①; Nozumi et al., 2017). EME depends on F-actin located in the Z-axis direction (see Igarashi et al., 2018). GPM6a (the symbol “m” in blue), distributed in the lipid rafts, is endocytosed through EME. The EME-dependent vesicles move in a retrograde direction to the central (C-) domain of the growth cone (②). Classical clathrin-mediated endocytosis (CME) mainly occurs at the bottom of the growth cone membrane (GCM; ③).
Several studies revealed that the impaired endocytosis of the lipid raft domains induced abnormal neuronal signaling, suggesting that lipid rafts are critical for endocytotic signaling pathways (Laudati et al., 2016; Nogueira-Rodrigues et al., 2016). Biochemically, these signaling events were thought to be clathrin-dependent (Qiu et al., 2011); however, super-resolution live-imaging of GPM6a- and cholesterol-dependent endocytosis in the growth cone revealed that these events were clathrin-independent (Nozumi et al., 2017) and dynamin and endophilin dependent. Dynamin is a GTPase that contributes to membrane cleavage and endocytosis (De Camilli et al., 1995). Endophilin is a BAR-domain protein that regulates membrane curvature (Kjaerulff et al., 2011; Gallop, 2020). The characteristics of these endocytotic events somewhat resemble “fast (or ultrafast) endocytosis,” a process seen at presynaptic terminals (Watanabe et al., 2014, 2018; Wu et al., 2014; Boucrot et al., 2015; Renard et al., 2015; Watanabe and Boucrot, 2017; Milosevic, 2018).
Phosphorylation at Stage 3 for Axon Growth
At stage 3, lipid rafts are thought to still be involved in axon formation via signaling in response to axon guidance molecules, including events such as protein phosphorylation (Guirland et al., 2004; Hérincs et al., 2005; Kamiguchi, 2006). It has been reported that cholesterol is more enriched in the growth cone at early stages than at later stages (Chauhan et al., 2020), suggesting the importance of the lipid rafts in this process. GAP-43 (growth-associated protein of 43-kDa), a neuronal growth-associated membrane protein also is known to be a lipid raft resident in the developing brain (Denny, 2006; Tong et al., 2008; Sekino-Suzuki et al., 2013; Kalinowska et al., 2015; Forsova and Zakharov, 2016). Recently, using phosphoproteomics of the growth cone, the most frequently phosphorylated site (among all of the identified growth cone membrane (GCM) proteins) was identified as S96 of GAP-43 (Kawasaki et al., 2018). The responsible kinase was identified as JNK, an enzyme whose activity also is dependent upon signaling in lipid rafts (Makdissy et al., 2018). Originally, JNK was postulated to be the transducer of apoptotic signals in multiple cell types (Hibi et al., 1993; Bogoyevitch et al., 2010); in neurons, this pathway was shown to induce axon degeneration (Shin et al., 2012). Three isoforms of the kinase (JNK1, 2, and 3) have been long been known to be related to cell death; recently, however, there is accumulating evidence that JNK has positive roles in neuronal development in the brain (Waetzig et al., 2006; Tararuk et al., 2006), including neurogenesis (Amura et al., 2005; Xu et al., 2014; Lim et al., 2015), neuronal migration (Kawauchi et al., 2003; Westerlund et al., 2011; Myers et al., 2014, 2020; Kawauchi, 2015), polarization (Slater et al., 2013), and axon growth and guidance (Oliva et al., 2006; Shafer et al., 2011; Feltrin et al., 2012; Qu et al., 2013; Sun et al., 2013). As has been hypothesized for other kinases (e.g., PKA, Akt, GSKβ, Cdk5, and Rho), JNK activation and phosphorylation of other substrates (Kawasaki et al., 2018; Ishikawa et al., 2019) is physiologically necessary for axon growth in the developing brain (Yamasaki et al., 2011). Protein phosphorylation is an important regulatory mechanism in cell development and homeostasis (Humphrey et al., 2015). At stage 3, rapid axon growth requires a signaling trigger, and protein phosphorylation is the most likely mediator of such a trigger.
Pin1 is a member of the peptidyl-prolyl isomerases (PPIases), a class of proteins that bind phosphorylated S/T-P motifs and catalyze the cis/trans-isomerization of P-containing peptides. This reaction switches the conformation and thereby the function(s) of the substrate proteins, including activity, protein-protein interaction, stability, and subcellular localization (Yaffe et al., 1997; Lu et al., 1999; Park et al., 2012; Litchfield et al., 2015). Pin1 is enriched in the brain and has been shown (by proteomics) to be present in the growth cone (Nozumi et al., 2009; Estrada-Bernal et al., 2012; Igarashi, 2014; Chauhan et al., 2020). The protein is known to activate protein kinases that participate in phosphorylation cascades (Litchfield et al., 2015). Indeed, JNK is directly kept activated by Pin1 (Park et al., 2012; Litchfield et al., 2015), suggesting that JNK activation via Pin1 likely occurs in the developing neuron and the growth cone from stages 1 to 3.
Recent advances have yielded phosphoproteomics, a powerful method for comprehensive and quantitative identification of the in vivo phosphorylation sites used in a given system (von Stechow et al., 2015; Invergo and Beltrao, 2018). Our application of this technique to GCM proteins (Ellis et al., 1985) led to the identification of more than 30,000 phosphopeptides representing ~4,600 different phosphorylation sites in ~1,200 proteins (Kawasaki et al., 2018; Igarashi et al., 2020). The phosphorylation of several frequently phosphorylated sites, including the S96 and T172 peptides of GAP-43 and the S25 and S1201 peptides of microtubule-associated protein 1B (MAP1B), is JNK dependent (Kawasaki et al., 2018; Ishikawa et al., 2019; Figure 4A). GAP-43 and MAP1B are classical axon growth markers and are highly phosphorylated (Skene, 1989; Riederer, 2007; Holahan, 2017). However, a subset of these frequently phosphorylated sites [e.g., peptide S41 of GAP-43, which is phosphorylated by PKC (Denny, 2006)] were not detected by phosphoproteomic analysis of the GCM. It remains unclear whether JNK is activated only in the cell bodies of the developing neuron, or in the axons or the growth cone at stage 3. If activation occurs in the cell bodies, JNK-phosphorylated substrates would have to undergo anterograde axonal transport to the growth cone; alternatively, if activation occurs in the axon or growth cone, JNK would catalyze local modification of substrates (Figure 4B).
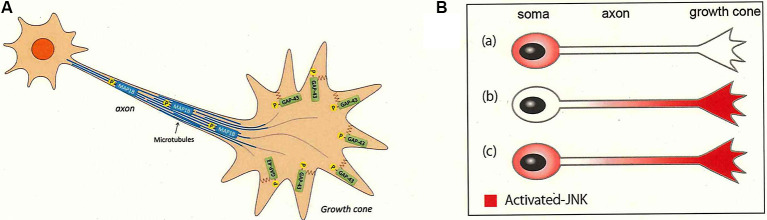
Figure 4.
JNK activity in the axon and its substrates for axonal growth. JNK is activated in the developing neurons (Hirai et al., 2011; Yamasaki et al., 2011; Coffey, 2014). (A) JNK-dependent substrates are sorted to the distal axon and the growth cone. Phosphorylated segments of GAP-43 (peptides pS96 and pT172) and MAP1B (peptides pS25 and pS1201) are sorted to the plasma membrane and the microtubules in the growth cone of the distal axon, respectively. These substrate proteins are phosphorylated by JNK in the cell bodies before undergoing anterograde axonal transport or are phosphorylated by JNK proximal to the growth cone area [see (A)]. See Kawasaki et al. (2018) and Ishikawa et al. (2019). (B) JNK may be distributed within the growing axons in one of three patterns: (Ba) only in the cell bodies, (Bb) only in the growth cone, or (Bc) in the whole neuron. Our experimental results indicate that (Ba) or (Bc) are more likely (Kawasaki et al., 2018).
Selected groups of C. elegans neurons have been found to require JNK and its upstream kinase (DLK, also referred to as MAP3K) for the regeneration of their axons (see review by Shimizu and Hisamoto, 2020); notably, a lack of JNK resulted in abnormal axonal growth (Tank et al., 2011). Thus, elevated JNK activity appears to be needed for axon maintenance in a wide range of organisms. However, the proteins located downstream of JNK in the C. elegans pathway (Chen et al., 2011) appeared to be totally different from the highly phosphorylated substrates identified in our phosphoproteomic analysis, and the JNK-dependent phosphorylated sites, which were analyzed using bioinformatic tools, appeared to be conserved only within components of the analogous vertebrate pathway (Igarashi and Okuda, 2019). Thus, while the need for JNK activity in axon growth/regeneration is conserved between model invertebrates and the mammalian central nervous system, JNK kinase appears to target distinct substrates and phosphorylation sites in these systems (Igarashi and Okuda, 2019).
Conclusions
In the context of mammalian neuronal polarization based on membrane trafficking, the molecular characterization of lipid rafts based on current detergent-resistant membrane fractions may not be sufficient to understand the hotspots of neuronal signaling. One of the new techniques addressing this issue is enzyme-mediated activation of the radical source (EMARS), which provides specific labeling of lipid rafts (Kotani et al., 2018). Given that only small amounts of proteins are collected after EMARS labeling, this method is still under development; nonetheless, the efficient nature of this labeling procedure holds promise for further expansion of its application.
We may re-examine the signaling pathways for polarity determination in neurons, each of which was previously examined by independent experiments. Portions of these pathways may be related to each other, and others may be proceeded independently and in parallel. Some of the earliest experiments are currently thought to be inappropriate for determining RNAi specificity, or for application to in vivo neuronal development of the brain. Also, in retrospect, several of the earlier experiments in this field would not have been able to discriminate effects in neuronal polarization from those in rapid axon growth. As mentioned above, the tug-of-war model developed based on in vitro results cannot simply be extended to in vivo polarization, given that neurons in vivo need to initiate growth in a synchronized fashion and to extend their axons in a single consistent direction.
Such re-examination also may require new methods. For example, phosphoproteomic analysis and super-resolution microscopy are expected to be powerful tools for characterizing the role of trafficking mechanisms in neuronal polarization. Although neuronal polarization and axonal growth appear to be relatively simple, understanding of these events has been more of a challenge than understanding other developmental stages; this difference may reflect the fact that considerably larger numbers of the proteins are involved in these processes, as proteomic and phosphoproteomic analyses have revealed.
Author Contributions
AH, AK, and MN produced the figures based upon their own original articles. MI wrote the article.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. Masayasu Okada, Yasuyuki Ito, Shujiro Okuda, and Atsushi Tamada for their support.
Footnotes
Funding. This work was supported in part by KAKENHI of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (#18H04013 and #18H04670 to MI, #15K06770 and 18K06480 to AH, #19K05947 to AK, and #15K06769 and #18K06459 to MN); by grants from the Takeda Science Foundation (to AH and MN); and by grants from AMED-CREST (Japan Agency for Medical Research and Development; #19gm1210007s0101 and #20gm1210007s0102 to MI) of the Agency for Medical Research and Development (AMED) of Japan.
References
- Aberg K. A., Dean B., Shabalin A. A., Chan R. F., Han L. K. M., Zhao M., et al. (2020). Methylome-wide association findings for major depressive disorder overlap in blood and brain and replicate in independent brain samples. Mol. Psychiatry 25, 1344–1354. 10.1038/s41380-018-0247-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo A., González-Billault C. (2018). Crosstalk between Rac1-mediated actin regulation and ROS production. Free Radic. Biol. Med. 116, 101–113. 10.1016/j.freeradbiomed.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Amura C. R., Marek L., Winn R. A., Heasley L. E. (2005). Inhibited neurogenesis in JNK1-deficient embryonic stem cells. Mol. Cell Biol. 25, 10791–10802. 10.1128/mcb.25.24.10791-10802.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelopoulou E., Paudel Y. N., Shaikh M. F., Piperi C. (2020). Flotillin: a promising biomarker for Alzheimer’s disease. J. Pers. Med. 10:E20. 10.3390/jpm10020020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaife-Lopes N., Sousa V. C., Pereira D. B., Ribeiro J. A., Chao M. V., Sebastião A. M. (2010). Activation of adenosine A2A receptors induces TrkB translocation and increases BDNF-mediated phospho-TrkB localization in lipid rafts: implications for neuromodulation. J. Neurosci. 30, 8468–8480. 10.1523/JNEUROSCI.5695-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Baumgart F., Corral-Escariz M., Pérez-Gil J., Rodríguez-Crespo I. (2010). Palmitoylation of R-Ras by human DHHC19, a palmitoyl transferase with a CaaX box. Biochim. Biophys. Acta 1798, 592–604. 10.1016/j.bbamem.2010.01.002 [DOI] [PubMed] [Google Scholar]
- Bhattacherjee A., Mu Y., Winter M. K., Knapp J. R., Eggimann L. S., Gunewardena S. S., et al. (2017). Neuronal cytoskeletal gene dysregulation and mechanical hypersensitivity in a rat model of Rett syndrome. Proc. Natl. Acad. Sci. U S A 114, E6952–E6961. 10.1016/j.bbamem.2010.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch M. A., Ngoei K. R., Zhao T. T., Yeap Y. Y., Ng D. C. (2010). c-Jun N-terminal kinase (JNK) signaling: recent advances and challenges. Biochim. Biophys. Acta 1804, 463–475. 10.1016/j.bbapap.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Boucrot E., Ferreira A. P., Almeida-Souza L., Debard S., Vallis Y., Howard G., et al. (2015). Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 517, 460–465. 10.1038/nature14067 [DOI] [PubMed] [Google Scholar]
- Brown D. A., London E. (1998). Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14, 111–136. 10.1146/annurev.cellbio.14.1.111 [DOI] [PubMed] [Google Scholar]
- Bruurs L. J., Bos J. L. (2014). Mechanisms of isoform specific Rap2 signaling during enterocytic brush border formation. PLoS One 9:e106687. 10.1371/journal.pone.0106687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M. R., Marta C. B., Kim F. S., Bansal R. (2009). Phosphorylation and lipid raft association of fibroblast growth factor receptor-2 in oligodendrocytes. Glia 57, 935–946. 10.1002/glia.20818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland S. L., Sanders S. S., Schmidt M. E., Riechers S. P., Lin D. T., Martin D. D., et al. (2014). The palmitoyl acyltransferase HIP14 shares a high proportion of interactors with huntingtin: implications for a role in the pathogenesis of Huntington’s disease. Hum. Mol. Genet. 23, 4142–4160. 10.1093/hmg/ddu137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracci M. O., Fuentealba L. M., Marzolo M. P. (2019). Golgi complex dynamics and its implication in prevalent neurological disorders. Front. Cell Dev. Biol. 7:75. 10.3389/fcell.2019.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan M. Z., Arcuri J., Park K. K., Zafar M. K., Fatmi R., Hackam A. S., et al. (2020). Multi-omic analyses of growth cones at different developmental stages provides insight into pathways in adult neuroregeneration. iScience 23:100836. 10.1016/j.isci.2020.100836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Gao J., Wu J., Zhang M., Cai M., Xu H., et al. (2015). Revealing the carbohydrate pattern on a cell surface by super-resolution imaging. Nanoscale 7, 3373–3380. 10.1039/c4nr05970k [DOI] [PubMed] [Google Scholar]
- Chen L., Wang Z., Ghosh-Roy A., Hubert T., Yan D., O’Rourke S., et al. (2011). Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron 71, 1043–1057. 10.1016/j.neuron.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini B., Parenti M. (2009). G-protein-coupled receptors, cholesterol, and palmitoylation: facts about fats. J. Mol. Endocrinol. 42, 371–379. 10.1677/jme-08-0114 [DOI] [PubMed] [Google Scholar]
- Coffey E. T. (2014). Nuclear and cytosolic JNK signalling in neurons. Nat. Rev. Neurosci. 15, 285–299. 10.1038/nrn3729 [DOI] [PubMed] [Google Scholar]
- Dang P., Smythe E., Furley A. J. W. (2012). TAG1 regulates the endocytic trafficking and signaling of the semaphorin3A receptor complex. J. Neurosci. 32, 10370–10382. 10.1523/JNEUROSCI.5874-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davare M. A., Fortin D. A., Saneyoshi T., Nygaard S., Kaech S., Banker G., et al. (2009). Transient receptor potential canonical 5 channels activate Ca2+/calmodulin kinase Iγ to promote axon formation in hippocampal neurons. J. Neurosci. 29, 9794–9808. 10.1523/JNEUROSCI.1544-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P., Takei K., McPherson P. S. (1995). The function of dynamin in endocytosis. Curr. Opin. Neurobiol. 5, 559–565. 10.1016/0959-4388(95)80059-x [DOI] [PubMed] [Google Scholar]
- Demmerle J., Innocent C., North A. J., Ball G., Müller M., Miron E., et al. (2017). Strategic and practical guidelines for successful structured illumination microscopy. Nat. Protoc. 12, 988–1010. 10.1038/nprot.2017.019 [DOI] [PubMed] [Google Scholar]
- Decker L., Baron W., Ffrench-Constant C. (2004). Lipid rafts: microenvironments for integrin-growth factor interactions in neural development. Biochem. Soc. Trans. 32, 426–430. 10.1042/bst0320426 [DOI] [PubMed] [Google Scholar]
- Denny J. B. (2006). Molecular mechanisms, biological actions, and neuropharmacology of the growth-associated protein GAP-43. Curr. Neuropharmacol. 4, 293–304. 10.2174/157015906778520782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti C. G., Sullivan C. A., Banker G. A. (1988). The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8, 1454–1468. 10.1523/JNEUROSCI.08-04-01454.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey P., Jorgenson K., Roy S. (2018). Actin assemblies in the axon shaft—some open questions. Curr. Opin. Neurobiol. 51, 163–167. 10.1016/j.conb.2018.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupraz S., Hilton B. J., Husch A., Santos T. E., Coles C. H., Stern S., et al. (2019). RhoA controls axon extension independent of specification in the developing brain. Curr. Biol. 29, 3874.e9–3886.e9. 10.1016/j.cub.2019.09.040 [DOI] [PubMed] [Google Scholar]
- Egawa J., Zemljic-Harpf A., Mandyam C. D., Niesman I. R., Lysenko L. V., Kleschevnikov A. M., et al. (2018). Neuron-targeted caveolin-1 promotes ultrastructural and functional hippocampal synaptic plasticity. Cereb. Cortex 28, 3255–3266. 10.1093/cercor/bhx196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis L., Wallis I., Abreu E., Pfenninger K. H. (1985). Nerve growth cones isolated from fetal rat brain. IV. Preparation of a membrane subfraction and identification of a membrane glycoprotein expressed on sprouting neurons. J. Cell Biol. 101, 1977–1989. 10.1083/jcb.101.5.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A. M., Toomre D., Bogan J. S. (2019). Acylation—a new means to control traffic through the golgi. Front. Cell Dev. Biol. 7:109. 10.3389/fcell.2019.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch T., Lemmon V., Banker G. (1999). Local presentation of substrate molecules directs axon specification by cultured hippocampal neurons. J. Neurosci. 19, 6417–6426. 10.1523/JNEUROSCI.19-15-06417.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Bernal A., Sanford S. D., Sosa L. J., Simon G. C., Hansen K. C., Pfenninger K. H. (2012). Functional complexity of the axonal growth cone: a proteomic analysis. PLoS One 7:e31858. 10.1371/journal.pone.0031858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltrin D., Fusco L., Witte H., Moretti F., Martin K., Letzelter M., et al. (2012). Growth cone MKK7 mRNA targeting regulates MAP1b-dependent microtubule bundling to control neurite elongation. PLoS Biol. 10:e1001439. 10.1371/journal.pbio.1001439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsova O. S., Zakharov V. V. (2016). High-order oligomers of intrinsically disordered brain proteins BASP1 and GAP-43 preserve the structural disorder. FEBS J. 283, 1550–1569. 10.1111/febs.13692 [DOI] [PubMed] [Google Scholar]
- Fujitani M., Honda A., Hata K., Yamagishi S., Tohyama M., Yamashita T. (2005). Biological activity of neurotrophins is dependent on recruitment of Rac1 to lipid rafts. Biochem. Biophys. Res. Commun. 327, 150–154. 10.1016/j.bbrc.2004.11.151 [DOI] [PubMed] [Google Scholar]
- Fukata Y., Murakami T., Yokoi N., Fukata M. (2016). Local palmitoylation cycles and specialized membrane domain organization. Curr. Top. Membr. 77, 97–141. 10.1016/bs.ctm.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Kobayashi H., Ishibashi K., Ohbayashi N. (2011). Genome-wide investigation of the Rab binding activity of RUN domains: development of a novel tool that specifically traps GTP-Rab35. Cell Struct. Funct. 36, 155–170. 10.1247/csf.11001 [DOI] [PubMed] [Google Scholar]
- Funahashi Y., Watanabe T., Kaibuchi K. (2020). Advances in defining signaling networks for the establishment of neuronal polarity. Curr. Opin. Cell Biol. 63, 76–87. 10.1016/j.ceb.2019.12.009 [DOI] [PubMed] [Google Scholar]
- Funk A. J., McCullumsmith R. E., Haroutunian V., Meador-Woodruff J. H. (2012). Abnormal activity of the MAPK- and cAMP-associated signaling pathways in frontal cortical areas in postmortem brain in schizophrenia. Neuropsychopharmacology 37, 896–905. 10.1038/npp.2011.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop J. L. (2020). Filopodia and their links with membrane traffic and cell adhesion. Semin. Cell Dev. Biol. 102, 81–89. [DOI] [PubMed] [Google Scholar]
- Gao L., Chen J., Gao J., Wang H., Xiong W. (2017). Super-resolution microscopy reveals the insulin-resistance-regulated reorganization of GLUT4 on plasma membranes. J. Cell Sci. 130, 396–405. 10.1242/jcs.192450 [DOI] [PubMed] [Google Scholar]
- Grider M. H., Park D., Spencer D. M., Shine H. D. (2009). Lipid raft-targeted Akt promotes axonal branching and growth cone expansion via mTOR and Rac1, respectively. J. Neurosci. Res. 87, 3033–3042. 10.1002/jnr.22140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirland C., Suzuki S., Kojima M., Lu B., Zheng J. Q. (2004). Lipid rafts mediate chemotropic guidance of nerve growth cones. Neuron 42, 51–62. 10.1016/s0896-6273(04)00157-6 [DOI] [PubMed] [Google Scholar]
- Guo C. L., Cheng P. L. (2015). Second messenger signaling for neuronal polarization: cell mechanics-dependent pattern formation. Dev. Neurobiol. 75, 388–401. 10.1002/dneu.22217 [DOI] [PubMed] [Google Scholar]
- Gustafsson M. G. (2008). Super-resolution light microscopy goes live. Nat. Methods 5, 385–387. 10.1038/nmeth0508-385 [DOI] [PubMed] [Google Scholar]
- Haack F., Lemcke H., Ewald R., Rharass T., Uhrmacher A. M. (2015). Spatio-temporal model of endogenous ROS and raft-dependent WNT/β-catenin signaling driving cell fate commitment in human neural progenitor cells. PLoS Comput. Biol. 11:e1004106. 10.1371/journal.pcbi.1004106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. M., Chu T. W., Peterson E. M., Zhang R., Yang J., Harris J., et al. (2015). Super-resolution imaging and quantitative analysis of membrane protein/lipid raft clustering mediated by cell-surface self-assembly of hybrid nanoconjugates. Chembiochem 16, 1725–1729. 10.1002/cbic.201500278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M., Wojcik M., Kim D., Mahmoudi M., Li W., Xu K. (2017). Correlative super-resolution microscopy: new dimensions and new opportunities. Chem. Rev. 117, 7428–7456. 10.1021/acs.chemrev.6b00604 [DOI] [PubMed] [Google Scholar]
- Hayashi T. (2020). Post-translational palmitoylation of ionotropic glutamate receptors in excitatory synaptic functions. Br. J. Pharmacol. [Epub ahead of print]. 10.1111/bph.15050 [DOI] [PubMed] [Google Scholar]
- Hérincs Z., Corset V., Cahuzac N., Furne C., Castellani V., Hueber A. O., et al. (2005). DCC association with lipid rafts is required for netrin-1-mediated axon guidance. J. Cell Sci. 118, 1687–1692. 10.1242/jcs.02296 [DOI] [PubMed] [Google Scholar]
- Hibi M., Lin A., Smeal T., Minden A., Karin M. (1993). Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 7, 2135–2148. 10.1101/gad.7.11.2135 [DOI] [PubMed] [Google Scholar]
- Hirai S., Banba Y., Satake T., Ohno S. (2011). Axon formation in neocortical neurons depends on stage-specific regulation of microtubule stability by the dual leucine zipper kinase-c-Jun N-terminal kinase pathway. J. Neurosci. 31, 6468–6480. 10.1523/JNEUROSCI.5038-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holahan M. R. (2017). A shift from a pivotal to supporting role for the growth-associated protein (GAP-43) in the coordination of axonal structural and functional plasticity. Front. Cell. Neurosci. 11:266. 10.3389/fncel.2017.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A., Ito Y., Takahashi-Niki K., Matsushita N., Nozumi M., Tabata H., et al. (2017a). Extracellular signals induce glycoprotein M6a clustering of lipid rafts and associated signaling molecules. J. Neurosci. 37, 4046–4064. 10.1523/JNEUROSCI.3319-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A., Usui H., Sakimura K., Igarashi M. (2017b). Rufy3 is an adapter protein for small GTPases that activates a Rac guanine nucleotide exchange factor to control neuronal polarity. J. Biol. Chem. 292, 20936–20946. 10.1074/jbc.m117.809541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey S. J., James D. E., Mann M. (2015). Protein phosphorylation: a major switch mechanism for metabolic regulation. Trends Endocrinol. Metab. 26, 676–687. 10.1016/j.tem.2015.09.013 [DOI] [PubMed] [Google Scholar]
- Igarashi M. (2014). Proteomic identification of the molecular basis of mammalian CNS growth cones. Neurosci. Res. 88, 1–15. 10.1016/j.neures.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Igarashi M. (2019). Molecular basis of the functions of the mammalian neuronal growth cone revealed using new methods. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 95, 358–377. 10.2183/pjab.95.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M., Kozaki S., Terakawa S., Kawano S., Ide C., Komiya Y. (1996). Growth cone collapse and inhibition of neurite growth by Botulinum neurotoxin C1: a t-SNARE is involved in axonal growth. J. Cell Biol. 134, 205–215. 10.1083/jcb.134.1.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M., Nozumi M., Wu L.-G., Cella Zanacchi F., Katona I., Barna L., et al. (2018). New observations in neuroscience using superresolution microscopy. J. Neurosci. 38, 9459–9467. 10.1523/JNEUROSCI.1678-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M., Okuda S. (2019). Evolutionary analysis of proline-directed phosphorylation sites in the mammalian growth cone identified using phosphoproteomics. Mol. Brain 12:53. 10.1186/s13041-019-0476-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M., Kawasaki A., Ishikawa Y., Honda A., Okada M., Okuda S. (2020). Phosphoproteomic and bioinformatic methods for analyzing signaling in vertebrate axon growth and regeneration. J. Neurosci. Methods 339:108723. 10.1016/j.jneumeth.2020.108723 [DOI] [PubMed] [Google Scholar]
- Invergo B. M., Beltrao P. (2018). Reconstructing phosphorylation signalling networks from quantitative phosphoproteomic data. Essays Biochem. 62, 525–534. 10.1042/ebc20180019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y., Okada M., Honda A., Ito Y., Tamada A., Endo N., et al. (2019). Phosphorylation sites of microtubule-associated protein 1B (MAP1B) involved in axon growth and regeneration. Mol. Brain 12:93. 10.1186/s13041-019-0510-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitsuka R., Saito T., Osada H., Ohno-Iwashita Y., Kobayashi T. (2011). Fluorescence image screening for chemical compounds modifying cholesterol metabolism and distribution. J. Lipid. Res. 52, 2084–2094. 10.1194/jlr.d018184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Honda A., Igarashi M. (2018). Glycoprotein M6a as a signaling transducer in neuronal lipid rafts. Neurosci. Res. 128, 19–24. 10.1016/j.neures.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Johnson D. M., Abi-Mansour J. P., Maurer J. A. (2012). Spatial confinement instigates environmental determination of neuronal polarity. Integr. Biol. 4, 1034–1037. 10.1039/c2ib20126g [DOI] [PubMed] [Google Scholar]
- Kalinowska M., Castillo C., Francesconi A. (2015). Quantitative profiling of brain lipid raft proteome in a mouse model of fragile X syndrome. PLoS One 10:e0121464. 10.1371/journal.pone.0121464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguchi H. (2006). The region-specific activities of lipid rafts during axon growth and guidance. J. Neurochem. 98, 330–335. 10.1111/j.1471-4159.2006.03888.x [DOI] [PubMed] [Google Scholar]
- Kanda A., Noda K., Yuki K., Ozawa Y., Furukawa T., Ichihara A., et al. (2013). Atp6ap2/(pro)renin receptor interacts with Par3 as a cell polarity determinant required for laminar formation during retinal development in mice. J. Neurosci. 33, 19341–19351. 10.1523/JNEUROSCI.1362-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., et al. (2008). Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909. 10.1038/nature07605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki A., Okada M., Tamada A., Okuda S., Nozumi M., Ito Y., et al. (2018). Growth cone phosphoproteomics reveals that GAP-43 phosphorylated by JNK is a marker of axon growth and regeneration. iScience 4, 190–203. 10.1016/j.isci.2018.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T. (2015). Cellullar insights into cerebral cortical development: focusing on the locomotion mode of neuronal migration. Front. Cell. Neurosci. 9:394. 10.3389/fncel.2015.00394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T., Chihama K., Nabeshima Y., Hoshino M. (2003). The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 22, 4190–4201. 10.1093/emboj/cdg413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaerulff O., Brodin L., Jung A. (2011). The structure and function of endophilin proteins. Cell Biochem. Biophys. 60, 137–154. 10.1007/s12013-010-9137-5 [DOI] [PubMed] [Google Scholar]
- Ko M., Zou K., Minagawa H., Yu W., Gong J. S., Yanagisawa K., et al. (2005). Cholesterol-mediated neurite outgrowth is differently regulated between cortical and hippocampal neurons. J. Biol. Chem. 280, 42759–42765. 10.1074/jbc.m509164200 [DOI] [PubMed] [Google Scholar]
- Köster M., Dell’Orco D., Koch K. W. (2014). The interaction network of rhodopsin involving the heterotrimeric G-protein transducin and the monomeric GTPase Rac1 is determined by distinct binding processes. FEBS J. 281, 5175–5185. 10.1111/febs.13064 [DOI] [PubMed] [Google Scholar]
- Kotani N., Nakano T., Ida Y., Ito R., Hashizume M., Yamaguchi A., et al. (2018). Analysis of lipid raft molecules in the living brain slices. Neurochem. Int. 119, 140–150. 10.1016/j.neuint.2017.08.012 [DOI] [PubMed] [Google Scholar]
- Kukimoto-Niino M., Takagi T., Akasaka R., Murayama K., Uchikubo-Kamo T., Terada T., et al. (2006). Crystal structure of the RUN domain of the RAP2-interacting protein x. J. Biol. Chem. 281, 31843–31853. 10.1074/jbc.M604960200 [DOI] [PubMed] [Google Scholar]
- Lalli G. (2014). Regulation of neuronal polarity. Exp. Cell Res. 328, 267–275. 10.1016/j.yexcr.2014.07.033 [DOI] [PubMed] [Google Scholar]
- Laudati E., Gilder A. S., Lam M. S., Misasi R., Sorice M., Gonias S. L., et al. (2016). The activities of LDL Receptor-related Protein-1 (LRP1) compartmentalize into distinct plasma membrane microdomains. Mol. Cell. Neurosci. 76, 42–51. 10.1016/j.mcn.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnerie C., Solecki D. J. (2018). Regulation of polarity protein levels in the developing central nervous system. J. Mol. Biol. 430, 3472–3480. 10.1016/j.jmb.2018.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledesma M. D., Simons K., Dotti C. G. (1998). Neuronal polarity: essential role of protein-lipid complexes in axonal sorting. Proc. Natl. Acad. Sci. U S A 95, 3966–3971. 10.1073/pnas.95.7.3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S., Lee S. J., Lee S. H., Ryu J. M., Lim H. S., Kim J. S., et al. (2016). Netrin-1-induced stem cell bioactivity contributes to the regeneration of injured tissues via the lipid raft-dependent integrin α6β4 signaling pathway. Sci. Rep. 6:37526. 10.1038/srep37526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim N. R., Yeap Y. Y., Zhao T. T., Yip Y. Y., Wong S. C., Xu D., et al. (2015). Opposing roles for JNK and Aurora A in regulating the association of WDR62 with spindle microtubules. J. Cell Sci. 128, 527–540. 10.1242/jcs.157537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder M. E., Deschenes R. J. (2007). Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84. 10.1038/nrm2084 [DOI] [PubMed] [Google Scholar]
- Lingwood D., Simons K. (2010). Lipid rafts as a membrane-organizing principle. Science 327, 46–50. 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- Litchfield D. W., Shilton B. H., Brandl C. J., Gyenis L. (2015). Pin1: intimate involvement with the regulatory protein kinase networks in the global phosphorylation landscape. Biochim. Biophys. Acta 1850, 2077–2086. 10.1016/j.bbagen.2015.02.018 [DOI] [PubMed] [Google Scholar]
- Lorent J. H., Levental I. (2015). Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem. Phys. Lipids 192, 23–32. 10.1016/j.chemphyslip.2015.07.022 [DOI] [PubMed] [Google Scholar]
- Lu P. J., Zhou X. Z., Shen M., Lu K. P. (1999). Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283, 1325–1328. 10.1126/science.283.5406.1325 [DOI] [PubMed] [Google Scholar]
- Ma C., Gu C., Huo Y., Li X., Luo X. J. (2018). The integrated landscape of causal genes and pathways in schizophrenia. Transl. Psychiatry 8:67. 10.1038/s41398-018-0114-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makdissy N., Haddad K., AlBacha J. D., Chaker D., Ismail B., Azar A., et al. (2018). Essential role of ATP6AP2 enrichment in caveolae/lipid raft microdomains for the induction of neuronal differentiation of stem cells. Stem Cell Res. Ther. 9:132. 10.1186/s13287-018-0862-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandyam C. D., Schilling J. M., Cui W., Egawa J., Niesman I. R., Kellerhals S. E., et al. (2017). Neuron-targeted caveolin-1 improves molecular signaling, plasticity and behavior dependent on the hippocampus in adult and aged mice. Biol. Psychiatry 81, 101–110. 10.1016/j.biopsych.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meka D. P., Scharrenberg R., Zhao B., Kobler O., König T., Schaefer I., et al. (2019). Radial somatic F-actin organization affects growth cone dynamics during early neuronal development. EMBO Rep. 20:e47743. 10.15252/embr.201947743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic I. (2018). Revisiting the role of clathrin-mediated endoytosis in synaptic vesicle recycling. Front. Cell. Neurosci. 12:27. 10.3389/fncel.2018.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möbius W., Patzig J., Nave K. A., Werner H. B. (2008). Phylogeny of proteolipid proteins: divergence, constraints, and the evolution of novel functions in myelination and neuroprotection. Neuron Glia Biol. 4, 111–127. 10.1017/s1740925x0900009x [DOI] [PubMed] [Google Scholar]
- Mori T., Wada T., Suzuki T., Kubota Y., Inagaki N. (2007). Singar1, a novel RUN domain-containing protein, suppresses formation of surplus axons for neuronal polarity. J. Biol. Chem. 282, 19884–19893. 10.1074/jbc.M700770200 [DOI] [PubMed] [Google Scholar]
- Myers A. K., Cunningham J. G., Smith S. E., Snow J. P., Smoot C. A., Tucker E. S. (2020). JNK signaling is required for proper tangential migration and laminar allocation of cortical interneurons. Development 147:dev180646. 10.1242/dev.180646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. K., Meechan D. W., Adney D. R., Tucker E. S. (2014). Cortical interneurons require Jnk1 to enter and navigate the developing cerebral cortex. J. Neurosci. 34, 7787–7801. 10.1523/JNEUROSCI.4695-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T., Kibe Y., Funahashi Y., Nakamuta S., Takano T., Ueno T., et al. (2014). Pioneering axons regulate neuronal polarization in the developing cerebral cortex. Neuron 81, 814–829. 10.1016/j.neuron.2013.12.015 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Yamaguchi T., Kato K., Yoshizawa M., Nabeshima Y., Ohno S., et al. (2005). PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat. Cell Biol. 7, 270–277. 10.1038/ncb1227 [DOI] [PubMed] [Google Scholar]
- Nogueira-Rodrigues J., Brites P., Sousa M. M. (2016). Axonal pathology in Krabbe’s disease: the cytoskeleton as an emerging therapeutic target. J. Neurosci. Res. 94, 1037–1041. 10.1002/jnr.23771 [DOI] [PubMed] [Google Scholar]
- Nozumi M., Igarashi M. (2018). Vesicular movements of the growth cone. Neurochem. Int. 119, 71–76. 10.1016/j.neuint.2017.09.011 [DOI] [PubMed] [Google Scholar]
- Nozumi M., Nakatsu F., Katoh K., Igarashi M. (2017). Coordinated movement of vesicles and actin bundles during nerve growth revealed by superresolution microscopy. Cell Rep. 18, 2203–2216. 10.1016/j.celrep.2017.02.008 [DOI] [PubMed] [Google Scholar]
- Nozumi M., Togano T., Takahashi-Niki K., Lu J., Honda A., Taoka M., et al. (2009). Identification of functional marker proteins in the mammalian growth cone. Proc. Natl. Acad. Sci. U S A 106, 17211–17216. 10.1073/pnas.0904092106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno-Iwashita Y., Shimada Y., Waheed A. A., Hayashi M., Inomata M., Nakamura M., et al. (2004). Perfringolysin O, a cholesterol-binding cytolysin, as a probe for lipid rafts. Anaerobe 10, 125–134. 10.1016/j.anaerobe.2003.09.003 [DOI] [PubMed] [Google Scholar]
- Oliva A. A., Jr., Atkins C. M., Copenagle L., Banker G. A. (2006). Activated c-Jun N-terminal kinase is required for axon formation. J. Neurosci. 26, 9462–9470. 10.1523/JNEUROSCI.2625-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. M., Magenau A., Williamson D., Gaus K. (2012). The lipid raft hypothesis revisited—new insights on raft composition and function from super-resolution fluorescence microscopy. Bioessays 34, 739–747. 10.1002/bies.201200044 [DOI] [PubMed] [Google Scholar]
- Park J. E., Lee J. A., Park S. G., Lee D. H., Kim S. J., Kim H. J., et al. (2012). A critical step for JNK activation: isomerization by the prolyl isomerase Pin1. Cell Death Differ. 19, 153–161. 10.1038/cdd.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Wang Y., Law P. Y., Chen H. Z., Loh H. H. (2011). Cholesterol regulates micro-opioid receptor-induced β-arrestin 2 translocation to membrane lipid rafts. Mol. Pharmacol. 80, 210–218. 10.1124/mol.110.070870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu C., Li W., Shao Q., Dwyer T., Huang H., Yang T., et al. (2013). c-Jun N-terminal kinase 1 (JNK1) is required for coordination of netrin signaling in axon guidance. J. Biol. Chem. 288, 1883–1895. 10.1074/jbc.m112.417881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan K., Kenworthy A. K. (2018). Dynamic pattern generation in cell membranes: current insights into membrane organization. Biochim. Biophys. Acta Biomembr. 1860, 2018–2031. 10.1016/j.bbamem.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randlett O., Poggi L., Zolessi F. R., Harris W. A. (2011). The oriented emergence of axons from retinal ganglion cells is directed by laminin contact in vivo. Neuron 70, 266–280. 10.1016/j.neuron.2011.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard H. F., Simunovic M., Lemière J., Boucrot E., Garcia-Castillo M. D., Arumugam S., et al. (2015). Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature 517, 493–496. 10.1038/nature14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh M. D. (2016). Fatty acylation of proteins: the long and the short of it. Prog. Lipid. Res. 63, 120–131. 10.1016/j.plipres.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K. N., Revelo N. H., Seitz K. J., Helm M. S., Sarkar D., Saleeb R. S., et al. (2018). Glyoxal as an alternative fixative to formaldehyde in immunostaining and super-resolution microscopy. EMBO J. 37, 139–159. 10.15252/embj.201695709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer B. M. (2007). Microtubule-associated protein 1B, a growth-associated and phosphorylated scaffold protein. Brain Res. Bull. 71, 541–558. 10.1016/j.brainresbull.2006.11.012 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Asiain A., Ruiz-Babot G., Romero W., Cubí R., Erazo T., Biondi R. M., et al. (2011). Brain specific kinase-1 BRSK1/SAD-B associates with lipid rafts: modulation of kinase activity by lipid environment. Biochim. Biophys. Acta 1811, 1124–1135. 10.1016/j.bbalip.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Saha S., Anilkumar A. A., Mayor S. (2016). GPI-anchored protein organization and dynamics at the cell surface. J. Lipid. Res. 57, 159–175. 10.1194/jlr.R062885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes D. H., Reh T. A., Harris W. A. (2019). Development of the Nervous System. 4th Edn. Academic Press: Elsevier. [Google Scholar]
- Schermelleh L., Ferrand A., Huser T., Eggeling C., Sauer M., Biehlmaier O., et al. (2019). Super-resolution microscopy demystified. Nat. Cell Biol. 21, 72–84. 10.1038/s41556-018-0251-8 [DOI] [PubMed] [Google Scholar]
- Schlegel J., Peters S., Doose S., Schubert-Unkmeir A., Sauer M. (2019). Super-resolution microscopy reveals local accumulation of plasma membrane gangliosides at Neisseria meningitidis invasion sites. Front. Cell Dev. Biol. 7:194. 10.3389/fcell.2019.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekino-Suzuki N., Yuyama K., Miki T., Kaneda M., Suzuki H., Yamamoto N., et al. (2013). Involvement of gangliosides in the process of Cbp/PAG phosphorylation by Lyn in developing cerebellar growth cones. J. Neurochem. 124, 514–522. 10.1111/jnc.12040 [DOI] [PubMed] [Google Scholar]
- Serjanov D., Bachay G., Hunter D. D., Brunken W. J. (2018). Laminin β2 chain regulates retinal progenitor cell mitotic spindle orientation via dystroglycan. J. Neurosci. 38, 5996–6010. 10.1523/JNEUROSCI.0551-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer B., Onishi K., Lo C., Colakoglu G., Zou Y. (2011). Vangl2 promotes wnt/planar cell polarity-like signaling by antagonizing Dvl1-mediated feedback inhibition in growth cone guidance. Dev. Cell 20, 177–191. 10.1016/j.devcel.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Hisamoto N. (2020). Factors regulating axon regeneration via JNK MAP kinase in C. elegans. J. Biochem. 167, 433–439. 10.1093/jb/mvaa020 [DOI] [PubMed] [Google Scholar]
- Shin J. E., Miller B. R., Babetto E., Cho Y., Sasaki Y., Qayum S., et al. (2012). SCG10 is a JNK target in the axonal degeneration pathway. Proc. Natl. Acad. Sci. U S A 109, E3696–E3705. 10.1073/pnas.1216204109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene J. H. (1989). Axonal growth-associated proteins. Annu. Rev. Neurosci. 12, 127–156. 10.1146/annurev.ne.12.030189.001015 [DOI] [PubMed] [Google Scholar]
- Slater P. G., Ramirez V. T., Gonzalez-Billault C., Varela-Nallar L., Inestrosa N. C. (2013). Frizzled-5 receptor is involved in neuronal polarity and morphogenesis of hippocampal neurons. PLoS One 8:e78892. 10.1371/journal.pone.0078892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley S. N., Warren M. F., Feldman R. J., Swerlick R. A., Mattheyses A. L., Kowalczyk A. P. (2016). Super-resolution microscopy reveals altered desmosomal protein organization in tissue from patients with pemphigus vulgaris. J. Invest. Dermatol. 136, 59–66. 10.1038/JID.2015.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanek O., Draber P., Horejsi V. (2014). Palmitoylated transmembrane adaptor proteins in leukocyte signaling. Cell. Signal. 26, 895–902. 10.1016/j.cellsig.2014.01.007 [DOI] [PubMed] [Google Scholar]
- Sui Z., Kovács A. D., Maggirwar S. B. (2006). Recruitment of active glycogen synthase kinase-3 into neuronal lipid rafts. Biochem. Biophys. Res. Commun. 345, 1643–1648. 10.1016/j.bbrc.2006.05.087 [DOI] [PubMed] [Google Scholar]
- Sun T., Yu N., Zhai L. K., Li N., Zhang C., Zhou L., et al. (2013). c-Jun NH2-terminal kinase (JNK)-interacting protein-3 (JIP3) regulates neuronal axon elongation in a kinesin- and JNK-dependent manner. J. Biol. Chem. 288, 14531–14543. 10.1074/jbc.m113.464453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sural-Fehr T., Singh H., Cantuti-Catelvetri L., Zhu H., Marshall M. S., Rebiai R., et al. (2019). Inhibition of the IGF-1-PI3K-Akt-mTORC2 pathway in lipid rafts increases neuronal vulnerability in a genetic lysosomal glycosphingolipidosis. Dis. Model. Mech. 12:dmm036590. 10.1242/dmm.036590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K. G. N., Ando H., Komura N., Fujiwara T. K., Kiso M., Kusumi A. (2017). Development of new ganglioside probes and unraveling of raft domain structure by single-molecule imaging. Biochim. Biophys. Acta. Gen. Subj. 1861, 2494–2506. 10.1016/j.bbagen.2017.07.012 [DOI] [PubMed] [Google Scholar]
- Takano T., Funahashi Y., Kaibuchi K. (2019). Neuronal polarity: positive and negative feedback signals. Front. Cell Dev. Biol. 7:69. 10.3389/fcell.2019.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada A., Igarashi M. (2017). Revealing chiral cell motility by 3D Riesz transform-differential interference contrast microscopy and computational kinematic analysis. Nat. Commun. 8:2194. 10.1038/s41467-017-02193-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank E. M., Rodgers K. E., Kenyon C. (2011). Spontaneous age-related neurite branching in Caenorhabditis elegans. J. Neurosci. 31, 9279–9288. 10.1523/JNEUROSCI.6606-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tararuk T., Ostman N., Li W., Björkblom B., Padzik A., Zdrojewska J., et al. (2006). JNK1 phosphorylation of SCG10 determines microtubule dynamics and axodendritic length. J. Cell Biol. 173, 265–277. 10.1083/jcb.200511055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin S. J., Cacao E. E., Hong D. W., Terenius L., Vukojevic V., Jovanovic-Talisman T. (2014). Nanoscale effects of ethanol and naltrexone on protein organization in the plasma membrane studied by photoactivated localization microscopy (PALM). PLoS One 9:e87225. 10.1371/journal.pone.0087225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J., Nguyen L., Vidal A., Simon S. A., Skene J. H., McIntosh T. J. (2008). Role of GAP-43 in sequestering phosphatidylinositol 4, 5-bisphosphate to raft bilayers. Biophys. J. 94, 125–133. 10.1529/biophysj.107.110536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa E., Hoogenraad C. C. (2018). Polarized trafficking: the palmitoylation cycle distributes cytoplasmic proteins to distinct neuronal compartments. Curr. Opin. Cell Biol. 50, 64–71. 10.1016/j.ceb.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Tortosa E., Adolfs Y., Fukata M., Pasterkamp R. J., Kapitein L. C., Hoogenraad C. C. (2017). Dynamic palmitoylation targets MAP6 to the axon to promote microtubule stabilization during neuronal polarization. Neuron 94, 809.e7–825.e7. 10.1016/j.neuron.2017.04.042 [DOI] [PubMed] [Google Scholar]
- Uechi Y., Bayarjargal M., Umikawa M., Oshiro M., Takei K., Yamashiro Y., et al. (2009). Rap2 function requires palmitoylation and recycling endosome localization. Biochem. Biophys. Res. Commun. 378, 732–737. 10.1016/j.bbrc.2008.11.107 [DOI] [PubMed] [Google Scholar]
- Vallejo D., Codocedo J. F., Inestrosa N. C. (2017). Posttranslational modifications regulate the postsynaptic localization of PSD-95. Mol. Neurobiol. 54, 1759–1776. 10.1007/s12035-016-9745-1 [DOI] [PubMed] [Google Scholar]
- Villarroel-Campos D., Bronfman F. C., Gonzalez-Billault C. (2016). Rab GTPase signaling in neurite outgrowth and axon specification. Cytoskeleton 73, 498–507. 10.1002/cm.21303 [DOI] [PubMed] [Google Scholar]
- von Stechow L., Francavilla C., Olsen J. V. (2015). Recent findings and technological advances in phosphoproteomics for cells and tissues. Expert Rev. Proteomics 12, 469–487. 10.1586/14789450.2015.1078730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waetzig V., Zhao Y., Herdegen T. (2006). The bright side of JNKs-Multitalented mediators in neuronal sprouting, brain development and nerve fiber regeneration. Prog. Neurobiol. 80, 84–97. 10.1016/j.pneurobio.2006.08.002 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Boucrot E. (2017). Fast and ultrafast endocytosis. Curr. Opin. Cell Biol. 47, 64–71. 10.1016/j.ceb.2017.02.013 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Mamer L. E., Raychaudhuri S., Luvsanjav D., Eisen J., Trimbuch T., et al. (2018). Synaptojanin and endophilin mediate neck formation during ultrafast endocytosis. Neuron 98, 1184.e6–1197.e6. 10.1016/j.neuron.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Trimbuch T., Camacho-Pérez M., Rost B. R., Brokowski B., Söhl-Kielczynski B., et al. (2014). Clathrin regenerates synaptic vesicles from endosomes. Nature 515, 228–233. 10.1038/nature13846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerlund N., Zdrojewska J., Padzik A., Komulainen E., Björkblom B., Rannikko E., et al. (2011). Phosphorylation of SCG10/stathmin-2 determines multipolar stage exit and neuronal migration rate. Nat. Neurosci. 14, 305–313. 10.1038/nn.2755 [DOI] [PubMed] [Google Scholar]
- Wu Y., O’Toole E. T., Girard M., Ritter B., Messa M., Liu X., et al. (2014). A dynamin 1-, dynamin 3- and clathrin-independent pathway of synaptic vesicle recycling mediated by bulk endocytosis. Elife 3:e01621. 10.7554/elife.01621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Xie G., Hu Y., Li X., Huang P., Zhang H. (2014). Neural differentiation of mesenchymal stem cells influences their chemotactic responses to stromal cell-derived factor-1α. Cell. Mol. Neurobiol. 34, 1047–1058. 10.1007/s10571-014-0082-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M. B., Schutkowski M., Shen M., Zhou X. Z., Stukenberg P. T., Rahfeld J. U., et al. (1997). Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278, 1957–1960. 10.1126/science.278.5345.1957 [DOI] [PubMed] [Google Scholar]
- Yamasaki T., Kawasaki H., Arakawa S., Shimizu K., Shimizu S., Reiner O., et al. (2011). Stress-activated protein kinase MKK7 regulates axon elongation in the developing cerebral cortex. J Neurosci. 31, 16872–16883. 10.1523/JNEUROSCI.1111-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang P., Wang G., Zhang H., Zhang Y., Yu Y., et al. (2018). Ras and Rap signal bidirectional synaptic plasticity via distinct subcellular microdomains. Neuron 98, 783.e4–800.e4. 10.1016/j.neuron.2018.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K., Xiang Y., Wang X., Wang Q., Zhong M., Wang S., et al. (2014). Epidermal growth factor receptor-PI3K signaling controls cofilin activity to facilitate herpes simplex virus 1 entry into neuronal cells. mBio 5:e00958–13. 10.1128/mbio.00958-13 [DOI] [PMC free article] [PubMed] [Google Scholar]