Abstract
Background: Recent studies have shown that blood-based miRNAs are dysregulated in patients with acute myocardial infarction (AMI) and are therefore a potential tool for the diagnosis of AMI. Therefore, this study summarized and evaluated studies focused on microRNAs as novel biomarkers for the diagnosis of AMI from the last ten years.
Methods: MEDLINE, the Cochrane Central database, and EMBASE were searched between January 2010 and December 2019. Studies that assessed the diagnostic accuracy of circulating microRNAs in AMI were chosen. The pooled sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, diagnostic odds ratio, and area under the curve (AUC) were used to assess the test performance of miRNAs.
Results: A total of 58 studies that included 8,206 participants assessed the diagnostic accuracy of circulating miRNAs in AMI. The main results of the meta-analyses are as follows: (1) Total miRNAs: the overall pooled sensitivity and specificity were 0.82 (95% CI: 0.79–0.85) and 0.87 (95% CI: 0.84–0.90), respectively. The AUC value was 0.91 (95% CI: 0.88–0.93) in the overall summary receiver operator characteristic (SROC) curve. (2) The panel of two miRNAs: sensitivity: 0.88 (95% CI: 0.77–0.94), specificity: 0.84 (95% CI: 0.72–0.91), AUC: 0.92 (95% CI: 0.90–0.94). (3) The panel of three miRNAs: sensitivity: 0.91 (95% CI: 0.85–0.94), specificity: 0.87 (95% CI: 0.77–0.92), AUC: 0.92 (95% CI: 0.89–0.94). (4) Results by types of miRNAs: miRNA-1: sensitivity: 0.78 (95% CI: 0.71–0.84), specificity: 0.86 (95% CI: 0.77–0.91), AUC: 0.88 (95% CI: 0.85–0.90); miRNA-133a: sensitivity: 0.85 (95% CI: 0.69–0.94), specificity: 0.92 (95% CI: 0.61–0.99), AUC: 0.93 (95% CI: 0.91–0.95); miRNA-208b: sensitivity: 0.80 (95% CI: 0.69–0.88), specificity: 0.96 (95% CI: 0.77–0.99), AUC: 0.91 (95% CI: 0.88–0.93); miRNA-499: sensitivity: 0.85 (95% CI: 0.77–0.91), specificity: 0.95 (95% CI: 0.89–0.98), AUC: 0.96 (95% CI: 0.94–0.97).
Conclusion: miRNAs may be used as potential biomarkers for the detection of AMI. For single, stand-alone miRNAs, miRNA-499 may have better diagnostic accuracy compared to other miRNAs. We propose that a panel of multiple miRNAs with high sensitivity and specificity should be tested.
Keywords: miRNAs, acute myocardial infarction, diagnosis, biomarker, meta-analysis
Introduction
Although advanced clinical medications have recently been developed for the diagnosis and prevention of coronary heart disease (CAD), acute myocardial infarction (AMI), which includes ST-elevation myocardial infarction (STEMI) and non-STEMI (NSTEMI), is still considered a primary public health threat, with high morbidity and mortality worldwide (Moss et al., 1996; GBD 2013 Mortality and Causes of Death Collaborators, 2015). Acute-phase reaction during ischemic damage is a crucial pathogenesis of ischemia myocardial issue (Hoffmeister et al., 2003). Dependent on accurate recognition and diagnosis, early effective revascularization treatment is an important strategy to repair ischemic myocardium and can significantly reduce the mortality of AMI patients (Hung et al., 2013). Currently, the most widely used biomarkers of myocardial injury during clinical practice are cardiac troponin and creatine kinase-MB (CK-MB), which may provide effective benefits for patients with revascularization therapy (Dohi et al., 2015; Anand et al., 2019). However, the elevation of cardiac troponin may be involved in serious, non-cardiac disease such as neuromuscular disorders, severe sepsis, and chronic renal insufficiency (Lamb et al., 2006; Finsterer et al., 2007; Vallabhajosyula et al., 2017). High levels of cardiac troponin have also been detected in patients with heart failure (Myhre et al., 2018). Therefore, early diagnostic biomarkers and improvement of the accuracy of approaches for the early prediction of AMI are still warranted.
Potential novel genetic and molecular biomarkers are currently being explored (Lorenzano et al., 2019). MicroRNAs (miRNAs/miRs) are endogenous, non-coding RNAs ~19–25nt that play crucial post-regulatory roles in animals and plants by targeting mRNAs for translational or cleavage repression (Bartel, 2004). MiRNAs can inhibit or reduce target gene expression, subsequently affecting protein expression (Saxena et al., 2018). Thus, miRNAs play important regulatory roles in cell growth, development, and differentiation (Gabisonia et al., 2019). Further, miRNAs have been identified in extracellular fluid and can be extremely stable despite the presence of endogenous RNase (Chevillet et al., 2014). In recent years, a number of studies have reported that miRNAs are dysregulated in CAD, and that specific circulating miRNA signatures might be useful as biomarkers for the diagnosis of AMI and as therapeutic targets (Jakob et al., 2017). However, the results of previous studies were significantly different, potentially due to sample size, specimen types, and different detection technologies. Therefore, the purpose of this systematic review and meta-analysis was to summarize the diagnostic values of blood-based miRNA levels from published articles from the last 10 years and to appraise the accuracy of results to determine whether miRNAs may be used as novel biomarkers for the diagnosis of AMI.
Materials and Methods
This study was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-analysis guidelines (Hutton et al., 2015; Moher et al., 2015). Two reviewers (CN Zhai, R Li) were independently involved with study selection, data extraction, and quality assessment.
Study Selection
An electronic search of MEDLINE (including PubMed), the Cochrane Central database, and EMBASE was performed to identify relevant articles published between January 2010 and December 2019. The following medical subject heading terms were used: (“plasma” OR “serum” OR “circulating”) AND (“microRNA” OR “miRNAs” OR “miR*”) AND (“myocardial infarction” OR “AMI” OR “coronary heart disease” OR “coronary artery disease” OR “coronary syndrome” OR “ischemic heart disease”). No language restrictions were imposed. All relevant review articles were retrieved, and duplicates were removed by manually searching. Based on the title and abstract, manuscripts of interest were obtained for full-text review. Only full-text references were included.
Inclusion and Exclusion Criteria
Inclusion and exclusion criteria were developed by the investigative team. The inclusion criteria were: (1) human studies, (2) studies related to circulating miRNAs levels and AMI, and (3) studies that contained enough data to evaluate the diagnostic value of miRNAs in AMI. Exclusion criteria were based on the following: (1) studies evaluating tissue miRNA or miRNA in other body fluids; (2) case reports, conference abstracts, and reviews; and (3) non-human studies.
Data Extraction, Meta-Analysis, and Quality Assessment
Each manuscript was assessed independently by two researchers (CN Zhai and K Hou). Disagreements among reviewers were resolved by consensus. Data extracted included the following: authors, publication year, country, type of blood-based fluid (serum or plasma), characteristics of the study population (both case and control), study design (qRT-PCR detection method), whether miRNA screening was performed, number of miRNAs assessed, listing of the specific dysregulated miRNAs in AMI patients compared with controls, and outcome of statistical analyses including details of miRNA analysis, such as type of reference miRNA utilized. Studies reporting on single miRNA were included in the meta-analysis and were evaluated according to the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) checklist (Whiting et al., 2011), which is designed to assess the risk of bias and the applicability of studies of diagnostic accuracy. The following four key domains were included: patient selection, the index test, the reference standard, and flow and timing. Each was assessed with respect to the risk of bias, and the first three domains were assessed with respect to applicability.
Statistical Analysis
Analysis was based on the accuracy of the identified miRNAs for diagnosing the presence of AMI, as determined using Receiver Operator Characteristic (ROC) curves via the Area Under the Curve (AUC) value, and sensitivity and specificity where available (Carter et al., 2016). We calculated the pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR), generated the bivariate summary receiver operator characteristic (SROC) curve, and calculated the area under the curve (AUC) to assess the overall diagnostic accuracy of miRNAs in distinguishing AMI patients from controls. Forest plots were constructed using STATA (15.0 StataCorp LP, College, Station, TX, USA). Due to the presumed heterogeneity of studies, a random-effects model (DerSimonian–Laird method) was used (Mahid et al., 2006). The heterogeneity of included studies was assessed using I2, and the P-value was considered significant if I2 was >50% or P < 0.05. Subgroup analyses were performed to explore the potential source of heterogeneity as follows: (1) based on the type of blood sample (plasma or serum), (2) the method of qRT-PCR detection (SYBR Green or TaqMan), (3) the type of reference control used for normalization (RNU or Cel-miRNA), (4) sample size (Sample size ≥ 100 or Sample size <100), and (5) different populations (Caucasian or East Asian). To assess the publication bias of the included studies, we performed Deeks' test of funnel plot asymmetry (Deeks et al., 2005).
Results
Literature Search Results and Characteristics of the Included Studies
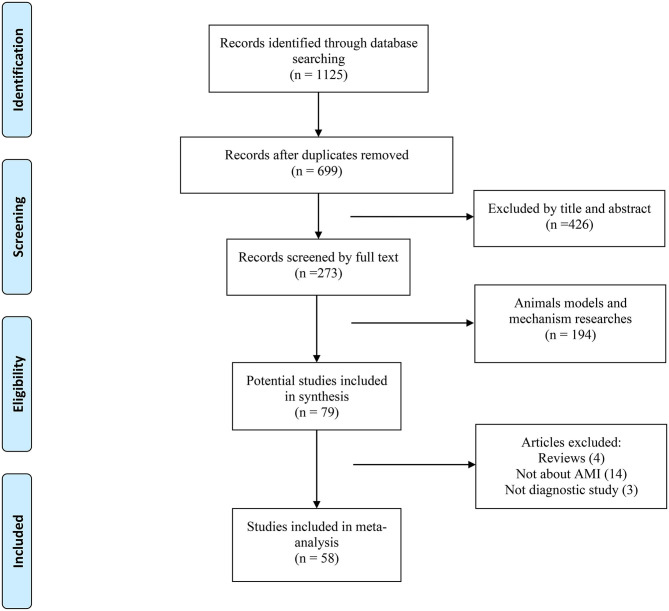
The PRISMA flow diagram of the literature search and inclusion of relevant studies are shown in Figure 1. Overall, 79 full-text articles were deemed relevant for a more detailed evaluation. Of these articles, 21 were excluded for the following reasons: review article (n = 4), not about AMI (n = 14), and not a diagnostic study (n = 3). Finally, 58 studies investigating plasma, serum, or peripheral venous blood miRNAs in the diagnosis of AMI were identified as eligible for inclusion in this systematic review (Adachi et al., 2010; Ai et al., 2010; Cheng et al., 2010; Corsten et al., 2010; D'Alessandra et al., 2010; Wang et al., 2010, 2011, 2013, 2014, 2016, 2019; Gidlöf et al., 2011, 2013; Meder et al., 2011; Zile et al., 2011; Devaux et al., 2012, 2015; Long et al., 2012a,b; Li C. et al., 2013; Li Y. Q. et al., 2013; Lu et al., 2013; Olivieri et al., 2013; He et al., 2014; Hsu et al., 2014; Huang et al., 2014; Li L. M. et al., 2014; Li Z. et al., 2014; Peng et al., 2014; Xiao et al., 2014; Białek et al., 2015; Chen et al., 2015; Gao et al., 2015; Han et al., 2015; Ji et al., 2015; Li et al., 2015; Liu et al., 2015, 2018; Yao et al., 2015; Zhang L. et al., 2015; Zhang R. et al., 2015; Zhao et al., 2015; Ke-Gang et al., 2016; Shalaby et al., 2016; Yang et al., 2016, 2017; Zhang et al., 2016, 2018; Zhu et al., 2016; Guo et al., 2017; Agiannitopoulos et al., 2018; Fawzy et al., 2018; Yi and An, 2018; Bukauskas et al., 2019; Li H. et al., 2019; Li P. et al., 2019; Xue et al., 2019a,b). These studies were performed in 12 countries; most of the subjects involved were East Asian, with Caucasian as the second most common population investigated. The major clinical characteristics of the included studies are shown in Table 1. In total, 8,206 patients were included in the study: 4,526 AMI patients and 3,680 healthy/non-AMI subjects (1,975 health controls, 1,705 non-AMI patients). Ten studies were relevant to the evaluation of patients with STEMI, three studies only included NSTEMI patients, and the other 45 articles included both types of myocardial infarction. The population demographics of our study are shown in Table 1. In total, 2,692 men and 1,834 women were included among AMI groups, and 2,066 men and 1,614 women were included in the control groups.
Figure 1.
Flow diagram of the literature search process and study inclusion.
Table 1.
Characteristics of studies included in the systematic review.
| References | Country | Specimen | Diseases | Case (n) | Control (n) | Age [mean ± s.d. /(range)] | Gender (male: female) | ||
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | ||||||
| Corsten et al. (2010) | Netherlands | Plasma | STEMI vs. NC | 32 | 36 | 62 ± 13 | 62 ± 13 | 24/12 | 23/13 |
| Adachi et al. (2010) | Japan | Plasma | AMI vs. NC | 9 | 10 | 66.8 ± 9.28 | 41.5 ± 8.0 | 6/3 | 5/5 |
| Ai et al. (2010) | China | Plasma | AMI vs. NC | 93 | 66 | 58.2 ± 10.2 | 55.1 ± 9.6 | 67/26 | 39/27 |
| Wang et al. (2010) | China | Plasma | AMI vs. non-AMI | 33 | 33 | 63.5 ± 10.1 | 64.3 ± 7.6 | 23/10 | 22/11 |
| Cheng et al. (2010) | China | Serum | AMI vs. NC | 31 | 20 | NA (45-71) | NA | 18/13 | NA |
| D'Alessandra et al. (2010) | Italy | Plasma | STEMI vs. NC | 33 | 17 | 57.9 ± 8.6 | 46.1 ± 13.9 | 31/2 | 13/4 |
| Gidlöf et al. (2011) | Sweden | Plasma | STEMI vs. NC | 25 | 11 | 64.56 ± 2.7 | 65.09 ± 3.51 | 20/5 | 7/4 |
| Meder et al. (2011) | Germany | Blood | AMI vs. NC | 20 | 20 | 59.3 ± 14 | 63.3 ± 14.8 | 16/4 | 14/6 |
| Wang et al. (2011) | China | Plasma | AMI vs. NC | 51 | 28 | 60.06 ± 11.53 | 57.86 ± 10.36 | 43/8 | 19/9 |
| Zile et al. (2011) | USA | Plasma | AMI vs. NC | 12 | 12 | 58 ± 3 | 61 ± 2 | 9/3 | 5/7 |
| Devaux et al. (2012) | Netherlands | Plasma | STEMI & NSTEMI vs. NC | 510 | 87 | 62 (0.32–91) | 53 (40–60) | 303/94 | 87/0 |
| Long et al. (2012a) | China | Plasma | AMI vs. NC | 17 | 25 | 53 ± 12.5 | 51 ± 12.3 | 13/4 | 18/7 |
| Long et al. (2012b) | China | Plasma | AMI vs. NC | 18 | 30 | 55 ± 11.4 | 50 ± 12.3 | 13/5 | 17/13 |
| Li Y. Q. et al. (2013) | China | Plasma | AMI vs. NC | 67 | 32 | 63.84 ± 11.17 | 61.75 ± 9.58 | 52/15 | 22/10 |
| Li C. et al. (2013) | China | Serum | AMI vs. NC | 117 | 100 | 62.7 ± 11.4 | 65.3 ± 9.98 | 20/97 | 19/81 |
| Lu et al. (2013) | China | Plasma | AMI vs. non-AMI | 40 | 15 | 66.8 ± 11.1 | 54.3 ± 17.5 | 30/10 | 6/9 |
| Gidlöf et al. (2013) | Sweden | Plasma | AMI vs. non-AMI | 319 | 88 | NA | NA | NA | NA |
| Olivieri et al. (2013) | Italy | Plasma | NSTEMI vs. NC | 92 | 99 | 82.6 ± 6.9 | 79.5 ± 5.4 | 39/53 | 40/59 |
| Wang et al. (2013) | China | Plasma | AMI vs. NC | 13 | 27 | NA | NA | NA | NA |
| Li L. M. et al. (2014) | China | Plasma | AMI vs. NC | 56 | 28 | 63.95 ± 11.34 | 60.50 ± 9.10 | 44/12 | 20/8 |
| Li Z. et al. (2014) | China | Plasma | AMI vs. NC | 27 | 31 | 54.15 ± 11.34 | 51.21 ± 12.25 | 20/7 | 16/15 |
| Huang et al. (2014) | China | Plasma | AMI vs. NC | 150 | 150 | 60.93 ± 12.86 | 60.66 ± 9.94 | 122/28 | 123/27 |
| Hsu et al. (2014) | Tai Wan | Serum | STEMI vs. NC | 31 | 31 | 59.0 ± 11.5 | 53.7 ± 14.8 | 29/2 | 29/2 |
| He et al. (2014) | China | Plasma | AMI vs. non-AMI | 359 | 30 | 58 ± 14 | 57 ± 10 | 301/58 | 21/9 |
| Peng et al. (2014) | China | Plasma | AMI vs. non-AMI | 76 | 110 | 64.6 (46–88) | 60 (52–81) | 43/33 | 61/49 |
| Wang et al. (2014) | China | Plasma | AMI vs. NC | 17 | 28 | 52 ± 11 | 58 ± 11 | 12/5 | 12/16 |
| Xiao et al. (2014) | China | Serum | AMI vs. NC | NA | NA | NA | NA | NA | NA |
| Białek et al. (2015) | Poland | Plasma | STEMI vs. non-AMI | 19 | 20 | 58 (55–65) | 63 (58–74) | 15/4 | 8/12 |
| Chen et al. (2015) | China | blood | AMI vs. non-AMI | 53 | 50 | 68.8 ± 7.3 | 68.3 ± 6.5 | 44/9 | 39/11 |
| Devaux et al. (2015) | Luxembourg | Plasma | AMI vs. non-AMI | 224 | 931 | 72 (61–80) | 61 (49–74) | 158/66 | 610/321 |
| Gao et al. (2015) | China | Plasma | STEMI vs. non-AMI | 35 | 160 | NA | NA | NA | NA |
| Ji et al. (2015) | China | Serum | AMI vs. NC | 98 | 23 | 64 ± 13.8 | 63.6 ± 12.2 | 82/16 | 15/8 |
| Han et al. (2015) | China | Plasma | AMI vs. non-AMI | 42 | 42 | 69.2 ± 7.4 | 67.4 ± 5.4 | 34/8 | 33/9 |
| Li et al. (2015) | China | Plasma | STEMI & NSTEMI Vs. NC | 87 | 87 | 56.93 ± 9.17 | 57.28 ± 10.82 | 64/23 | 62/25 |
| Liu et al. (2015) | China | Plasma | AMI vs. NC | 70 | 72 | 64.2 ± 11.2 | 62.3 ± 10.3 | 34/36 | 35/37 |
| Yao et al. (2015) | China | Plasma | AMI vs. NC | 50 | 39 | 63.2 ± 11.4 | 62.7 ± 10.5 | 33/17 | 27/12 |
| Zhang L. et al. (2015) | China | Plasma | AMI vs. non-AMI | 142 | 85 | 64.86 ± 12.84 | 66.45 ± 10.61 | 102/40 | 59/26 |
| Zhang R. et al. (2015) | China | Plasma | STEMI & NSTEMI | 110 | 110 | 57.74 ± 12.03 | 58.28 ± 11.32 | 87/23 | 83/27 |
| Zhao et al. (2015) | China | Serum | AMI vs. NC | 59 | 60 | 60.1 ± 11.3 | 61.9 ± 12.1 | 34/25 | 30/30 |
| Ke-Gang et al. (2016) | China | Plasma | AMI vs. non-AMI | 233 | 79 | 63 (20-91) | 69 (35-70) | 163/70 | 46/33 |
| Wang et al. (2016) | China | Plasma | AMI vs. NC | 32 | 36 | 55.62 ± 9.17 | 58.57 ± 11.54 | 18/14 | 24/12 |
| Yang et al. (2016) | China | Plasma | AMI vs. NC | 54 | 30 | NA | NA | NA | NA |
| Zhang et al. (2016) | China | Plasma | AMI vs. NC | 17 | 10 | 59.7 ± 8.4 | 56.2 ± 5.5 | 12/5 | 5/5 |
| Zhu et al. (2016) | China | Plasma | STEMI vs. NC | 60 | 60 | 64.1 ± 10.3 | 62.1 ± 11.2 | 46/14 | 45/15 |
| Shalaby et al. (2016) | Egypt | Serum | NSTEMI vs. NC | 48 | 25 | 54.3 ± 8.3 | 49.2 ± 10.2 | 32/16 | 17/8 |
| Fawzy et al. (2018) | Egypt | Serum | STEMI vs. NC | 110 | 121 | NA | NA | 61/49 | 63/58 |
| Guo et al. (2017) | China | Plasma | AMI vs. NC | 90 | 45 | 35.2 ± 5.6 | 36.2 ± 5.2 | 48/42 | 25/20 |
| Yang et al. (2017) | China | Serum | AMI vs. NC | 76 | 30 | 63.6 ± 11.7 | 62.3 ± 10.5 | 48/28 | 19/11 |
| Agiannitopoulos et al. (2018) | Greece | Plasma | AMI vs. NC | 50 | 50 | 62.12 ± 10.99 | 59.30 ± 9.82 | 38/12 | 33/17 |
| Liu et al. (2018) | China | Plasma | NSTEMI vs. NC | 145 | 30 | 67 | 65 | 56:89 | 15/15 |
| Wang et al. (2019) | China | Plasma | AMI vs. NC | 66 | 70 | 61.84 ± 1.71 | 61.90 ± 3.69 | 30/36 | 26/44 |
| Yi and An (2018) | China | Plasma | AMI vs. NC | 30 | 30 | 61.35 ± 8.65 | 57.64 ± 5.91 | NA | NA |
| Zhang et al. (2018) | China | Plasma | STEMI vs. NC | 80 | 60 | 62.83 ± 7.52 | 63.15 ± 7.32 | 46/34 | 35/25 |
| Bukauskas et al. (2019) | Lithuania | Serum | STEMI vs. NC | 62 | 26 | 64 ± 12 | 42 ± 13 | 46/16 | 20/6 |
| Li H. et al. (2019) | China | Plasma | AMI vs. NC | 35 | 55 | 60.86 ± 11.25 | 56.36 ± 12.36 | 15/20 | 27/28 |
| Li P. et al. (2019) | China | Serum | AMI vs. non-AMI | 41 | 32 | 62.95 ± 11.04 | 63.16 ± 10.63 | 30/11 | 19/13 |
| Xue et al. (2019a) | China | Plasma | STEMI & NSTEMI | 29 | 21 | 68.0 ± 10.4 | 58.5 ± 14.3 | 23/6 | 16/5 |
| Xue et al. (2019b) | China | Plasma | STEMI & NSTEMI | 31 | 27 | 61.1 ± 10.0 | 60.1 ± 12.2 | 25/6 | 19/8 |
STEMI, ST-elevation myocardial infarction; AMI, acute myocardial infarction; NSTEMI, non ST-elevation myocardial infarction; NC, normal control.
Identification of Dysregulated miRNAs in the Included Studies
All included studies used Quantitative reverse transcription polymerase chain reaction (qRT-PCR) to detect the expression levels of miRNAs. A summary of all study methods is provided in Table 2. Seven studies performed miRNA screening to compare blood-based miRNAs between AMI patients and control groups (Adachi et al., 2010; D'Alessandra et al., 2010; Meder et al., 2011; Li C. et al., 2013; Hsu et al., 2014; Huang et al., 2014; Li et al., 2015). Fifty-one articles identified miRNAs based on their own previous studies or based on the literature (Ai et al., 2010; Cheng et al., 2010; Corsten et al., 2010; Wang et al., 2010, 2011, 2013, 2014, 2016, 2019; Gidlöf et al., 2011, 2013; Zile et al., 2011; Devaux et al., 2012, 2015; Long et al., 2012a,b; Li Y. Q. et al., 2013; Lu et al., 2013; Olivieri et al., 2013; He et al., 2014; Li L. M. et al., 2014; Li Z. et al., 2014; Peng et al., 2014; Xiao et al., 2014; Białek et al., 2015; Chen et al., 2015; Gao et al., 2015; Han et al., 2015; Ji et al., 2015; Liu et al., 2015, 2018; Yao et al., 2015; Zhang L. et al., 2015; Zhang R. et al., 2015; Ke-Gang et al., 2016; Shalaby et al., 2016; Yang et al., 2016, 2017; Zhang et al., 2016, 2018; Zhu et al., 2016; Guo et al., 2017; Agiannitopoulos et al., 2018; Fawzy et al., 2018; Yi and An, 2018; Bukauskas et al., 2019; Li H. et al., 2019; Li P. et al., 2019; Xue et al., 2019a,b). The expression of 50 miRNAs were identified as either significantly higher or lower expression in AMI cases; specific details were shown in Table 3. Thirty-three miRNAs were upregulated (miRNA-1, miRNA-17-5p, miRNA-19b-3p, miRNA-21, miRNA-23b, miRNA-26a-1, miRNA-30a, miRNA-122-5p, miRNA-124, miRNA-126, miRNA-133a/b, miRNA-134, miRNA-145-3p, miRNA-146a, miRNA-150, miRNA-181a, miRNA-186, miRNA-195, miRNA-199a-1, miRNA-208a/b, miRNA-210, miRNA-223, miRNA-302b, miRNA-328, miRNA-361-5p, miRNA-423-5p, miRNA-486, miRNA-494, miRNA-497, miRNA-499, miRNA-663b, miRNA-1291, and miRNA-1303) (Adachi et al., 2010; Ai et al., 2010; Cheng et al., 2010; Corsten et al., 2010; D'Alessandra et al., 2010; Wang et al., 2010, 2011, 2013, 2014, 2016, 2019; Gidlöf et al., 2011, 2013; Meder et al., 2011; Zile et al., 2011; Devaux et al., 2012, 2015; Long et al., 2012a,b; Li C. et al., 2013; Li Y. Q. et al., 2013; Olivieri et al., 2013; He et al., 2014; Hsu et al., 2014; Li L. M. et al., 2014; Li Z. et al., 2014; Xiao et al., 2014; Białek et al., 2015; Chen et al., 2015; Han et al., 2015; Ji et al., 2015; Li et al., 2015; Liu et al., 2015, 2018; Yao et al., 2015; Zhang L. et al., 2015; Zhang R. et al., 2015; Ke-Gang et al., 2016; Shalaby et al., 2016; Zhang et al., 2016, 2018; Guo et al., 2017; Yang et al., 2017; Agiannitopoulos et al., 2018; Fawzy et al., 2018; Li P. et al., 2019; Xue et al., 2019a,b), and 17 miRNAs were downregulated (miRNA-22-5p, miRNA-23a-3p, miRNA-26a, miRNA-30d-5p, miRNA-99a, miRNA-125b, miRNA-126-3p, miRNA-132-5p, miRNA-145, miRNA-146a-5p, miRNA-191, miRNA-214, miRNA-320b, miRNA-375, miRNA-379, miRNA-519-5p, and miRNA-let-7d) (D'Alessandra et al., 2010; Meder et al., 2011; Long et al., 2012b; Lu et al., 2013; Hsu et al., 2014; Huang et al., 2014; Wang et al., 2014, 2019; Gao et al., 2015; Li et al., 2015; Yang et al., 2016; Yi and An, 2018; Bukauskas et al., 2019; Li H. et al., 2019). Thirteen upregulated miRNAs (miRNA-1, miRNA-19b-3p, miRNA-21, miRNA-122-5p, miRNA-126, miRNA-133a/b, miRNA-134, miRNA-150, miRNA-186, miRNA-208a/b, miRNA-486, miRNA-499, and miRNA-663b) (Adachi et al., 2010; Corsten et al., 2010; D'Alessandra et al., 2010; Wang et al., 2010, 2011, 2013, 2016, 2019; Gidlöf et al., 2011, 2013; Devaux et al., 2012, 2015; Li Y. Q. et al., 2013; Olivieri et al., 2013; He et al., 2014; Hsu et al., 2014; Peng et al., 2014; Xiao et al., 2014; Białek et al., 2015; Chen et al., 2015; Han et al., 2015; Ji et al., 2015; Li et al., 2015; Liu et al., 2015, 2018; Zhang L. et al., 2015; Zhang R. et al., 2015; Ke-Gang et al., 2016; Shalaby et al., 2016; Agiannitopoulos et al., 2018; Fawzy et al., 2018; Li H. et al., 2019; Li P. et al., 2019) and three downregulated miRNAs (miRNA-26a, miRNA-191, and miRNA-375) (D'Alessandra et al., 2010; Hsu et al., 2014; Li et al., 2015; Wang et al., 2019) were identified by more than one study. Among these articles, which included 22 original studies, miRNA-499 was the most frequently identified dysregulated miRNA in AMI patients (Adachi et al., 2010; Corsten et al., 2010; D'Alessandra et al., 2010; Wang et al., 2010; Gidlöf et al., 2011, 2013; Devaux et al., 2012, 2015; Li C. et al., 2013; Li Y. Q. et al., 2013; Olivieri et al., 2013; Xiao et al., 2014; Chen et al., 2015; Liu et al., 2015, 2018; Zhang L. et al., 2015; Zhang R. et al., 2015; Shalaby et al., 2016; Agiannitopoulos et al., 2018; Fawzy et al., 2018; Li P. et al., 2019). 16 studies focused on the miRNA-208 family (miRNA-208a/b) (Corsten et al., 2010; Wang et al., 2010; Gidlöf et al., 2011, 2013; Devaux et al., 2012, 2015; Li C. et al., 2013; Li Y. Q. et al., 2013; Xiao et al., 2014; Białek et al., 2015; Han et al., 2015; Li et al., 2015; Liu et al., 2015, 2018; Agiannitopoulos et al., 2018; Li P. et al., 2019), 14 studies focused on miRNA-1 (Ai et al., 2010; Cheng et al., 2010; Corsten et al., 2010; D'Alessandra et al., 2010; Wang et al., 2010; Gidlöf et al., 2011, 2013; Long et al., 2012a; Li C. et al., 2013; Li Y. Q. et al., 2013; Olivieri et al., 2013; Li L. M. et al., 2014; Liu et al., 2015, 2018), and 14 studies focused on the miRNA-133 family (miRNA-133a/b) (Corsten et al., 2010; D'Alessandra et al., 2010; Wang et al., 2010, 2011, 2013; Gidlöf et al., 2011; Li Y. Q. et al., 2013; Olivieri et al., 2013; Peng et al., 2014; Ji et al., 2015; Ke-Gang et al., 2016; Liu et al., 2018). Additionally, four studies identified a panel of two miRNAs (Hsu et al., 2014; Zhang R. et al., 2015; Shalaby et al., 2016; Wang et al., 2019), six studies identified a panel of three miRNAs (Long et al., 2012b; Wang et al., 2014, 2016; Li H. et al., 2019; Xue et al., 2019a,b), and one study identified a panel of more than four miRNAs that were elevated in AMI (Li C. et al., 2013).
Table 2.
Study methods and corresponding dysregulated miRNAs identified.
| References | qRT-PCR detection method | Reference control | miRNA screening performed | Dysregulated miRNAsa (n) | Significantly dysregulated miRNAs on validation (n) | AMI definition | Time of blood sampling |
|---|---|---|---|---|---|---|---|
| Corsten et al. (2010) | SYBR | n/a | N | 6 | 2 | ST segment elevation; increase CK and TnI | within 12 h of chest pain onset |
| Adachi et al. (2010) | TaqMan | n/a | Y | 3 | 1 | NA | Within 48 h after onset of chest pain |
| Ai et al. (2010) | SYBR | RNU6 | N | 2 | 1 | Ischemic symptoms; increase cTnI and CK-MB; ST segment elevation or depression; pathological Q wave | NA |
| Wang et al. (2010) | TaqMan | Cel-miR-39 | N | 40 | 4 | Ischemia symptom; increase cTnI; ST segment change; coronary angiography | Within 12 h after admission |
| Cheng et al. (2010) | n/a | n/a | N | 1 | 1 | Ischemic chest pain; increase CK-MB; ST segment elevation | Within 24 h after AMI |
| D'Alessandra et al. (2010) | TaqMan | n/a | Y | 48 | 6 | NA | Within 12 h after the onset of symptoms |
| Gidlöf et al. (2011) | SYBR | miR-16 | N | 5 | 4 | ST-segment elevation; coronary angiography | Within 24 h of the onset of ischemic symptoms |
| Meder et al. (2011) | n/a | RNU6 | Y | 40 | 20 | ESC/AHA guidelines | NA |
| Wang et al. (2011) | SYBR | RNU6 | N | 2 | 2 | Chest pain lasting >20 min; increase CK-MB and cTnI; ST segment and T- wave changes; pathological Q wave | Within 24 h after the onset of syndromes |
| Zile et al. (2011) | TaqMan | RNU6 | N | 6 | 5 | AHA/ACC guidelines | Within 24 h after admission |
| Devaux et al. (2012) | TaqMan | n/a | N | 2 | 2 | ST segment elevation or depression; increase CK and TnI; coronary angiography | With acute and ongoing chest pain for 12 h |
| Long et al. (2012a) | SYBR | RNU6 | N | 2 | 2 | Ischemic symptom; increase cTnI and CK-MB; ST segment elevation or depression; pathological Q wave; | Within 4 h after onset of symptom |
| Long et al. (2012b) | SYBR | RNU6 | N | 3 | 3 | Ischemic symptom; increase cTnI and CK-MB; ST segment elevation or depression; pathological Q wave; | Within 4 h after onset of symptom |
| Li Y. Q. et al. (2013) | SYBR | Cel-miR-39 | N | 4 | 4 | Chest pain lasting >30 min; increase CK-MB and cTnI; ST segment elevation or depression; pathological Q wave | Within 12 h of the onset of symptoms |
| Li C. et al. (2013) | TaqMan | n/a | Y | 21 | 6 | Chest pain; ST segment elevation or depression; increase cTnI and CK-MB; pathological Q wave | Within 2h after hospitalization |
| Lu et al. (2013) | TaqMan | RNU6 | N | 1 | 1 | Typical chest pain; increase cTnI; coronary angiography | The next morning after admission |
| Gidlöf et al. (2013) | SYBR | miR-17 | N | 3 | 2 | STEMI: ECG criteria; NSTEMI: increase troponin and clinical symptoms | 71% were taken within 24 h, 82% within 48 h and 93% within 72 h after onset of chest pain |
| Olivieri et al. (2013) | TaqMan | miR-17, cel-miR-39 | N | 6 | 5 | Ischemic symptom; ST segment elevation or depression >1 mm/negative T wave/new onset LVBB; increase cTnT | Immediately after hospitalization |
| Wang et al. (2013) | SYBR | RNU6 | N | 1 | 1 | Acute ischemic chest pain within 24 h; ECG changes; increase cTnI | Immediately after admission |
| Li L. M. et al. (2014) | SYBR | Cel-miR-39 | N | 1 | 1 | Increased cTnT or CK-MB; chest pain lasting for >30 min; pathological Q waves/ST-segment changes | Within 12 h after onset of chest pain |
| Li Z. et al. (2014) | SYBR | RNU6 | N | 1 | 1 | Ischemic symptoms; increase cTnI and CK-MB; ST segment elevation or depression; pathological Q wave | Within 4 h after onset of symptom |
| Huang et al. (2014) | n/a | Cel-miR-39 | Y | 77 | 2 | Chest paining lasting >20 min; ST segment changes; pathological Q wave; increase cardiac biomarkers | NA |
| Hsu et al. (2014) | SYBR | n/a | Y | 25 | 5 | ACC/AHA guideline | NA |
| He et al. (2014) | SYBR | n/a | N | 2 | 2 | Ischemic symptoms; increase cTn and CK; ST segment change; pathological Q wave | 6 h after the onset of symptoms |
| Peng et al. (2014) | TaqMan | miR-16 | N | 3 | 3 | STEMI: ST segment elevation; NSTEMI: ischemic symptom and increase cTnI | Within 3 h after admission |
| Wang et al. (2014) | SYBR | RNU6 | N | 3 | 3 | Acute ischemic-type chest pain; ECG changes; increase cTnI | Immediately after admission |
| Xiao et al. (2014) | TaqMan | Cel-miR-39 | N | 2 | 2 | NA | NA |
| Białek et al. (2015) | TaqMan | HY3 | N | 1 | 1 | Chest pain; ST segment elevation; coronary angiography | NA |
| Chen et al. (2015) | TaqMan | RNU6 | N | 1 | 1 | Biochemical markers; acute ischemic-type chest pain; ECG changes; coronary angiography | Immediately after admission |
| Devaux et al. (2015) | SYBR | n/a | N | 6 | 3 | ACC/AHA guideline | With acute chest pain for 12 h |
| Gao et al. (2015) | SYBR | Cel-miR-39 | N | 1 | 1 | ACC/AHA guidelines | Immediately after hospitalization |
| Ji et al. (2015) | SYBR | miR-16 | N | 3 | 3 | Ischemia symptoms; ST segment elevation; increase cTnI and CK-MB | Immediately after hospitalization |
| Han et al. (2015) | TaqMan | RNU6 | N | 1 | 1 | ESC/AHA/ACC guidelines | Within 12 h after the symptom onset |
| Li et al. (2015) | TaqMan | RNU6 | Y | 28 | 3 | Ischemia symptoms; ST segment abnormality; pathological Q wave; increase cTnI and CK-MB | Within 4 h after onset of symptoms |
| Liu et al. (2015) | TaqMan | Cel-miR-39 | N | 3 | 3 | Increased cTnT or CK-MB; chest pain lasting for >30 min; pathological Q waves/ST-segment changes | Within 2 h after the onset of symptom |
| Yao et al. (2015) | SYBR | RNU6 | N | 1 | 1 | Ischemic symptoms; increase cTnI and CK-MB; ST segment changes and pathological Q wave | Within 4 h after the onset of symptoms |
| Zhang L. et al. (2015) | TaqMan | RNU6 | N | 1 | 1 | Ischemic-type chest pain; increase cTnI; ECG change; coronary angiography | Within 12 h after the onset of acute chest pain |
| Zhang R. et al. (2015) | TaqMan | RNU6 | N | 2 | 2 | Acute ischemic chest pain; abnormal ECG; increase cTnI and CK | Within 24 h after admission |
| Zhao et al. (2015) | SYBR | Cel-miR-39 | N | 1 | 1 | ESC/ACC guidelines | Within 24 h after hospitalization |
| Ke-Gang et al. (2016) | TaqMan | Cel-miR-39 | N | 1 | 1 | ESC Guidelines | Immediately after hospitalization |
| Wang et al. (2016) | SYBR | Cel-miR-39 | N | 3 | 3 | ESC/AHA/ACC guidelines | Within 12 h after the onset of chest pain |
| Yang et al. (2016) | SYBR | RNU6 | N | 1 | 1 | Coronary angiography | NA |
| Zhang et al. (2016) | TaqMan | Cel-miR-39 | N | 1 | 1 | ESC/ACC guidelines | Immediately after admission |
| Zhu et al. (2016) | TaqMan | RNU6 | N | 1 | 1 | Ischemic chest pain lasting>30 min; increase cTnI; new ST segment elevation | Immediately after admission |
| Shalaby et al. (2016) | SYBR | RNU6 | N | 2 | 2 | Ischemic symptom; no ST segment elevation; increase cTnI; coronary angiography | Within 24 h of onset of chest pain |
| Fawzy et al. (2018) | TaqMan | RNU6 | N | 1 | 1 | Ischemic symptoms; ECG changes; coronary angiography | Within the first 12 h of the chest pain |
| Guo et al. (2017) | n/a | GAPDH | N | 1 | 1 | Guideline for AMI by Chinese Medicine Academy | Within 12 h of chest pain onset |
| Yang et al. (2017) | SYBR | RNU6 | N | 1 | 1 | Clinical symptoms; ECG changes; increase cTnI, CK and CK-MB | Within 24 h of symptoms onset |
| Agiannitopoulos et al. (2018) | TaqMan | RNU24 | N | 2 | 2 | Ischemic chest pain; ST segment elevation or depression; pathological Q wave; rise of cardiac biomarkers | Within 24 h of onset of chest pain |
| Liu et al. (2018) | SYBR | n/a | N | 4 | 4 | ACC guideline | Within 2–4 h after admission |
| Wang et al. (2019) | n/a | Cel-miR-39 | N | 6 | 2 | Ischemic symptoms; ST segment-T wave changes or new LBBB; pathological Q wave; coronary angiography | Within 4 h of onset of chest pain/dyspnea |
| Yi and An (2018) | SYBR | RNU6 | N | 1 | 1 | Coronary angiography; increase cTns and CK-MB | NA |
| Zhang et al. (2018) | SYBR | n/a | N | 1 | 1 | ST segment elevation; coronary angiography | Within 6 h after the onset of symptoms |
| Bukauskas et al. (2019) | SYBR | Cel-miR-39 | N | 3 | 3 | 2015 ESC Guidelines for the management of ACS | the first 24 h of admission |
| Li H. et al. (2019) | SYBR | Cel-miR-39 | N | 3 | 3 | ACC/AHA guideline | With chest pain for 10 h |
| Li P. et al. (2019) | SYBR | RNU6 | N | 4 | 4 | Ischemic symptoms; ST segment elevation or pathological Q wave; increase cTnI | With chest pain for 3 h |
| Xue et al. (2019a) | SYBR | Cel-miR-39 | N | 3 | 3 | 2012 ESC/AHA/ACC guidelines | Within 4 h of onset of chest pain |
| Xue et al. (2019b) | SYBR | Cel-miR-39 | N | 6 | 3 | 2012 ESC/AHA/ACC guidelines | Chest pain onset <4 h duration |
SYBR, SYBR Green; RNU6, small nuclear RNA U6; STEMI, ST-elevation myocardial infarction; NSTEMI, non ST-elevation myocardial infarction; CK, creatine kinase; CK-MB, creatine kinase-MB. Cardiac troponin I/T, cTnI/T. ECG, electrocardiogram.
(n): the number of dysregulated miRNAs.
Based upon miRNA screening, or based upon literature review or prior work (see text).
Y or N, Yes or No; NA: not available.
Table 3.
Significantly dysregulated miRNAs of patients with AMI as compared with controls.
| miRNAs | References | Expression | Area under the curve (AUC) | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Single miRNA | |||||
| miRNA-1 | Ai et al., 2010 Corsten et al., 2010 Cheng et al., 2010 D'Alessandra et al., 2010 Wang et al., 2010 Gidlöf et al., 2011 Li Y. Q. et al., 2013 Long et al., 2012a Olivieri et al., 2013 Li C. et al., 2013 Gidlöf et al., 2013 Li L. M. et al., 2014 Liu et al., 2015 Liu et al., 2018 |
Upregulated | 0.774 —a — — 0.847 0.979 0.827 0.92 — — 0.59 0.854 0.81 0.773 |
70.0% 90.0% — — 78.0% 88.9% 80.0% 93.0% — 78.0% 55.0% 80.0% 70.0% 75.7% |
75.0% 95.0% — — 80.0% 100% 90.0% 90.0% — 86.0% 65.0% 90.0% 90.0% 53.5% |
| miRNA-17-5p | Xue et al., 2019a | Upregulated | 0.857 | 85.2% | 85.7% |
| miRNA-19b-3p | Wang et al., 2016 Wang et al., 2019 |
Upregulated | 0.821 0.667 |
72.2% 66.1% |
66.7% 61.3% |
| miRNA-21 miRNA-21-5p |
Zile et al., 2011 Olivieri et al., 2013 Zhang et al., 2016 Wang et al., 2014 |
Upregulated | — — 0.892 0.949 |
— — 78.6% — |
— — 100% — |
| miRNA-23b | Zhang et al., 2018 | Upregulated | 0.809 | 88.1% | 60.3% |
| miRNA-26a-1 | Xue et al., 2019b | Upregulated | 0.965 | 100% | 85.2% |
| miRNA-30a | Long et al., 2012b | Upregulated | 0.88 | 88.3% | 82.5% |
| miRNA-122-5p | Yao et al., 2015 Wang et al., 2019 |
Upregulated | 0.855 0.626 |
100% 63.8% |
60.2% 73.9% |
| miRNA-124 | Guo et al., 2017 | Upregulated | 0.86 | 52.0% | 91.0% |
| miRNA-126 miRNA-126-5p |
Long et al., 2012a Xue et al., 2019a |
Upregulated | 0.86 0.802 |
75.1% 100% |
83.1% 61.9% |
| miRNA-133 miRNA-133a miRNA-133b |
Wang et al., 2011 Peng et al., 2014 Liu et al., 2018 Corsten et al., 2010 D'Alessandra et al., 2010 Wang et al., 2010 Gidlöf et al., 2011 Li Y. Q. et al., 2013 Olivieri et al., 2013 Wang et al., 2013 Ji et al., 2015 Ke-Gang et al., 2016 D'Alessandra et al., 2010 Ji et al., 2015 |
Upregulated | 0.89 0.912 0.928 — — 0.867 0.859 0.947 — — 0.787 0.667 — 0.823 |
98.0% 81.1% 71.0% — — 87.3% 99.6% 82.7% — — 85.4% 61% — 66.5% |
73.0% 91.2% 96.5% — — 84.9% 63.6% 100% — — 99.8% 68% — 100% |
| miRNA-134 miRNA-134-5p |
He et al., 2014 Li C. et al., 2013 Wang et al., 2016 Wang et al., 2019 |
Upregulated | 0.818 0.657 0.827 0.702 |
79.4% 53.6% 77.8% 70.6% |
77.1% 78.1% 77.8% 79.1% |
| miRNA-145-3p | Xue et al., 2019a | Upregulated | 0.720 | 81.8% | 61.9% |
| miRNA-146a | Xue et al., 2019b | Upregulated | 0.911 | 100% | 66.7% |
| miRNA-150-3p miRNA-150 |
Hsu et al., 2014 Li H. et al., 2019 Zhang R. et al., 2015 |
Upregulated | 0.715 0.904 0.678 |
71.3% 88.2% 80.6% |
70.1% 75.9% 49.8% |
| miRNA-181a | Zhu et al., 2016 | Upregulated | 0.834 | 81.5% | 81.8% |
| miRNA-186 miRNA-186-5p |
Li C. et al., 2013 Wang et al., 2016 Wang et al., 2019 |
Upregulated | 0.715 0.824 0.692 |
77.8% 77.8% 56.9% |
60.9% 77.8% 84.3% |
| miRNA-195 | Long et al., 2012b | Upregulated | 0.89 | 100% | 44.7% |
| miRNA-199a-1 | Xue et al., 2019b | Upregulated | 0.855 | 96.8% | 66.7% |
| miRNA-208b miRNA-208a miRNA-208 |
Corsten et al., 2010 Gidlöf et al., 2011 Devaux et al., 2012 Li Y. Q. et al., 2013 Gidlöf et al., 2013 Devaux et al., 2015 Li et al., 2015 Agiannitopoulos et al., 2018 Wang et al., 2010 Xiao et al., 2014 Białek et al. (2015) Li C. et al., 2013 Han et al., 2015 Liu et al., 2015 Liu et al., 2018 Li P. et al., 2019 |
Upregulated | 0.944 1.0 0.90 0.89 0.82 0.76 0.674 0.999 0.965 — — 0.778 — 0.72 0.9940 0.868 |
90.6% 100% 79.5% 82.4% 79.0% 64.7% 59.8% 98% 90.9% — — 75.8% — 65.0% 90.0% 70.0% |
94.1% 100% 99.8% 99.8% 70.0% 80.2% 73.6% 100% 100% — — 73.1% — 90.0% 100% 97.5% |
| miRNA-210 | Shalaby et al., 2016 | Upregulated | 0.90 | 83.3% | 100% |
| miRNA-223 | Li C. et al., 2013 | Upregulated | 0.741 | 77.8% | 68.3% |
| miRNA-302b | Yang et al., 2017 | Upregulated | 0.95 | 88.2% | 93.3% |
| miRNA-328 | He et al., 2014 | Upregulated | 0.887 | 86.3% | 74.6% |
| miRNA-361-5p | Wang et al., 2014 | Upregulated | 0.881 | — | — |
| miRNA-423-5p | Olivieri et al., 2013 | Upregulated | — | — | — |
| miRNA-486-3p miRNA-486 |
Hsu et al., 2014 Zhang R. et al., 2015 |
Upregulated | 0.629 0.731 |
38.8% 56.5% |
84.1% 86.5% |
| miRNA-494 | Li P. et al., 2019 | Upregulated | 0.839 | 79.6% | 82.1% |
| miRNA-497 | Li Z. et al., 2014 | Upregulated | 0.88 | 82% | 94% |
| miRNA-499 miRNA-499-5p miRNA-499a |
Corsten et al., 2010 Adachi et al., 2010 Wang et al., 2010 Devaux et al., 2012 Li Y. Q. et al., 2013 Li C. et al., 2013 Xiao et al., 2014 Chen et al., 2015 Devaux et al., 2015 Liu et al., 2015 Zhang L. et al., 2015 Zhao et al., 2015 Shalaby et al., 2016 Agiannitopoulos et al., 2018 Liu et al., 2018 Li P. et al., 2019 D'Alessandra et al., 2010 Gidlöf et al., 2011 Olivieri et al., 2013 Gidlöf et al., 2013 Fawzy et al., 2018 |
Upregulated | 0.918 — 0.822 0.98 0.884 0.755 — — 0.65 0.88 0.86 0.915 0.97 0.999 0.995 0.852 — 0.989 0.88 0.79 0.953 |
84.5% — 60.0% 95.0% 80.0% 80.0% — — 35.7% 82.0% 80% 86.37% 93.4% 98% 98.4% 71.5% — 87.7% 88.0% 64.0% 97.2% |
93.9% — 90.3% 100% 94.0% 93.0% — — 90.3% 94.0% 80.28% 93.47% 100% 100% 100% 89.3% — 100% 75.0% 90.0% 75% |
| miRNA-663b | Meder et al., 2011 Peng et al., 2014 |
Upregulated | 0.94 0.611 |
90% 72.4% |
95% 76.5% |
| miRNA-1291 | Peng et al., 2014 | Upregulated | 0.695 | 78.4% | 89.5% |
| miRNA-1303 | Li H. et al., 2019 | Upregulated | 0.884 | 81.2% | 89.3% |
| miRNA-22-5p | Wang et al., 2019 | Downregulated | 0.975 | 96.7% | 96.7% |
| miRNA-23a-3p | Bukauskas et al., 2019 | Downregulated | 0.806 | 73.3% | 79.6% |
| miRNA-26a-5p miRNA-26a |
Hsu et al., 2014 Li et al., 2015 |
Downregulated | 0.675 0.745 |
57.8% 73.6% |
90.2% 72.4% |
| miRNA-30d-5p | Bukauskas et al., 2019 | Downregulated | 0.745 | 80.9% | 64.9% |
| miRNA-99a | Yang et al., 2016 | Downregulated | — | — | — |
| miRNA-125b | Huang et al., 2014 | Downregulated | 0.858 | — | — |
| miRNA-126-3p | Hsu et al., 2014 | Downregulated | 0.694 | 64.1% | 80.0% |
| miRNA-132-5p | Li H. et al., 2019 | Downregulated | 0.886 | 85.3% | 74.1% |
| miRNA-145 | Gao et al., 2015 | Downregulated | — | — | — |
| miRNA-146a-5p | Bukauskas et al., 2019 | Downregulated | 0.800 | 84.5% | 70.4% |
| miRNA-191-5p miRNA-191 |
Hsu et al., 2014 Li et al., 2015 |
Downregulated | 0.652 0.669 |
48.6% 62.1% |
93.8% 69.0% |
| miRNA-214 | Lu et al., 2013 | Downregulated | — | — | — |
| miRNA-320b | Huang et al., 2014 | Downregulated | 0.866 | — | — |
| miRNA-375 | D'Alessandra et al., 2010 Wang et al., 2019 |
Downregulated | — 0.510 |
— 92.6% |
— 33.1% |
| miRNA-379 | Yi and An, 2018 | Downregulated | 0.751 | — | — |
| miRNA-519-5p | Wang et al., 2014 | Downregulated | 0.798 | — | — |
| miRNA-let-7d | Long et al., 2012b | Downregulated | 0.86 | 64.7% | 100% |
| Two miRNA panels | |||||
| miRNA-191-5p,-486-3p | Hsu et al., 2014 | Upregulated and downregulatedb | 0.867 | 83.87% | 83.33% |
| miRNA-486-3p,-126-3p | Hsu et al., 2014 | Upregulated | 0.849 | 61.29% | 93.55% |
| miRNA-126-3p,-150-3p | Hsu et al., 2014 | Upregulated | 0.843 | 93.55% | 64.52% |
| miRNA-486-3p,-26a-5p | Hsu et al., 2014 | Upregulated and downregulated | 0.821 | 87.10% | 64.52% |
| miRNA-26a-5p,-150-3p | Hsu et al., 2014 | Upregulated and downregulated | 0.821 | 83.87% | 70.97% |
| miRNA-191-5p,-150-3p | Hsu et al., 2014 | Upregulated and downregulated | 0.789 | 77.42% | 77.42% |
| miRNA-150,-486 | Zhang R. et al., 2015 | Upregulated | 0.771 | 72.6% | 72.1% |
| miRNA-499,-210 | Shalaby et al., 2016 | Upregulated | 0.98 | 97.9% | 100% |
| miRNA-22-5p,-122-5p | Wang et al., 2019 | Upregulated and downregulated | 0.976 | 98.4% | 96.2% |
| Three miRNA panels | |||||
| miRNA-30a,-195, let-7d | Long et al., 2012b | Upregulated and downregulated | 0.930 | 94% | 90% |
| miRNA-21-5p,-361-5p,-519-5p | Wang et al., 2014 | Upregulated and downregulated | 0.989 | 93.9% | 100% |
| miRNA-19b-3p,-134-5p,-186-5p | Wang et al., 2016 | Upregulated | 0.898 | 88.9% | 77.8% |
| miRNA-22-5p,-150-3p,-132-5p | Li H. et al., 2019 | Upregulated and downregulated | 0.942 | 91.2% | 87.0% |
| miRNA-17-5p,-126-5p,-145-3p | Xue et al., 2019a | Upregulated | 0.857 | 84.0% | 85.7% |
| miRNA-26a-1,-146a,-199a-1 | Xue et al., 2019b | Upregulated | 0.913 | 97.8% | 71.6% |
| Panels with ≥ 4 miRNAs | |||||
| miRNA-1,-134,-186,-208,-223,-499 | Li C. et al., 2013 | Upregulated | 0.811 | 55.3% | 90.1% |
AMI, acute myocardial infarction; miRNA, microRNA.
Not reported.
Upregulated and downregulated: where the individual miRNAs were either upregulated or downregulated in AMI compared with control groups.
Sensitivity, Specificity, and Area Under the Curve
Among the included studies, the most common methods of assessing the diagnostic accuracy of dysregulated miRNAs were AUC and sensitivity and specificity, as determined from ROC curves. In the identified dysregulated miRNAs, AUC values ranged from 0.510 to 1.0, sensitivity ranged from 48.6% to 100%, and specificity from 33.1% to 100%. In the single identified miRNAs, the highest AUC sensitivity and specificity combination was reported for miRNA-208b (AUC 1.0, sensitivity 100%, specificity 100%) (Gidlöf et al., 2011). In the panel of two miRNAs, the highest value combination was reposted for miRNA-499 and miRNA-210 (AUC 0.98, sensitivity 97.9%, specificity 100%) (Shalaby et al., 2016). In the panel of three miRNAs, the highest value combination was reported for miRNA-21-5p, miRNA-361-5p, and miRNA-519-5p (AUC 0.989, sensitivity 93.9%, specificity 100%) (Wang et al., 2014). In the panel of ≥ 4 miRNAs, there was one study about AUC 0.811, which had sensitivity 55.3% and specificity 90.1% (Li C. et al., 2013).
Meta-Analysis Outcomes of Diagnostic Accuracy of miRNAs in AMI
The Quality Assessment of Included Studies
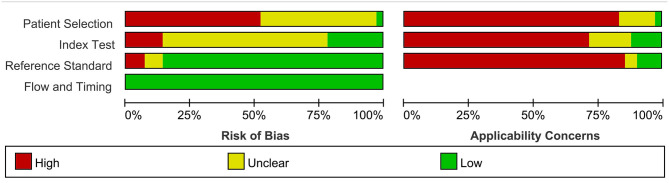
The pooled result of the meta-analysis for the diagnostic accuracy of blood-based miRNAs in AMI was pooled from 42 studies (Ai et al., 2010; Corsten et al., 2010; Wang et al., 2010, 2011, 2014, 2016, 2019; Gidlöf et al., 2011, 2013; Meder et al., 2011; Devaux et al., 2012, 2015; Long et al., 2012a,b; Li C. et al., 2013; Li Y. Q. et al., 2013; Olivieri et al., 2013; He et al., 2014; Hsu et al., 2014; Li L. M. et al., 2014; Li Z. et al., 2014; Peng et al., 2014; Ji et al., 2015; Li et al., 2015; Liu et al., 2015, 2018; Yao et al., 2015; Zhang L. et al., 2015; Zhang R. et al., 2015; Ke-Gang et al., 2016; Shalaby et al., 2016; Yang et al., 2016; Zhang et al., 2016, 2018; Zhu et al., 2016; Guo et al., 2017; Agiannitopoulos et al., 2018; Fawzy et al., 2018; Bukauskas et al., 2019; Li H. et al., 2019; Li P. et al., 2019; Xue et al., 2019a,b). Quality assessment results of the studies reporting on miRNAs included in the meta-analysis using the QUADAS-2 evaluation tool are shown in Supplementary Figure 1. Results are presented as percentages across the studies (Figure 2).
Figure 2.
Bar graphs of the methodological quality assessment. Each risk-of-bias and applicability item is presented as percentages across included studies, which indicates the proportion of different levels for each item.
Total miRNAs
Dysregulated miRNAs in AMI patients compared with controls, the SROC curve with AUC, sensitivity, specificity, PLR, NLR, and DOR for miRNAs were included in our meta-analysis (Table 4). A random effect model was used for the meta-analysis due to significant heterogeneity (all I2 > 50%). Forty-four individual miRNAs were identified in 43 studies (Ai et al., 2010; Corsten et al., 2010; Wang et al., 2010, 2011, 2014, 2016, 2019; Gidlöf et al., 2011, 2013; Meder et al., 2011; Devaux et al., 2012, 2015; Long et al., 2012a,b; Li C. et al., 2013; Li Y. Q. et al., 2013; Olivieri et al., 2013; He et al., 2014; Hsu et al., 2014; Li L. M. et al., 2014; Li Z. et al., 2014; Peng et al., 2014; Ji et al., 2015; Li et al., 2015; Liu et al., 2015, 2018; Yao et al., 2015; Zhang L. et al., 2015; Zhang R. et al., 2015; Ke-Gang et al., 2016; Shalaby et al., 2016; Yang et al., 2016; Zhang et al., 2016, 2018; Zhu et al., 2016; Guo et al., 2017; Agiannitopoulos et al., 2018; Fawzy et al., 2018; Bukauskas et al., 2019; Li H. et al., 2019; Li P. et al., 2019; Xue et al., 2019a,b). The sensitivity, specificity, and the corresponding SROC value with 95% CIs (95% confidential intervals) of the total miRNAs in the diagnostic of AMI were 0.82 (95% CI: 0.79–0.85), 0.87 (95 %CI: 0.84–0.90), and 0.91 (95% CI: 0.88–0.93), respectively (Supplementary Figures 2A,B, 3A). The Deeks' funnel plot asymmetry test suggested a potential for publication bias in the total miRNAs (p-value = 0.00, Supplementary Figure 3B).
Table 4.
The overall and subgroups meta-analysis results for comparison of diagnostic value of miRNAs.
| Comparisons (n, study) | AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) |
|---|---|---|---|---|---|---|
| POOLED SINGLE miRNAs (n = 102) | ||||||
| Total miRNAs | 0.91 (0.88–0.93) | 0.82 (0.79–0.85) | 0.87 (0.84–0.90) | 6.27 (4.97–7.90) | 0.21 (0.18–0.24) | 30.40 (21.60–42.77) |
| Multiple combinations | ||||||
| Two miRNAs panel (n = 9) | 0.92 (0.90–0.94) | 0.88 (0.77–0.94) | 0.84 (0.72–0.91) | 5.40 (2.84–10.26) | 0.15 (0.07–0.30) | 37.00 (10.52–130.16) |
| Three miRNAs panel (n = 6) | 0.92 (0.89–0.94) | 0.91 (0.85–0.94) | 0.87 (0.77–0.92) | 6.70 (3.83–11.72) | 0.11 (0.07–0.18) | 62.24 (27.40–141.38) |
| SUBGROUP ANALYSIS | ||||||
| Type of blood sample | ||||||
| Plasma (n = 75) | 0.92 (0.89–0.94) | 0.84 (0.80–0.87) | 0.87 (0.82–0.90) | 6.22 (4.68–8.27) | 0.19 (0.15–0.23) | 32.97 (21.48–50.61) |
| Serum (n = 26) | 0.89 (0.86–0.92) | 0.78 (0.72–0.82) | 0.87 (0.82–0.91) | 6.13 (4.23–8.89) | 0.26 (0.20–0.33) | 23.88 (14.15–40.28) |
| Type of miRNA detection method | ||||||
| SYBR green (n = 62) | 0.92 (0.89–0.94) | 0.84 (0.80–0.87) | 0.87 (0.83–0.90) | 6.33 (4.77–8.40) | 0.19 (0.15–0.24) | 33.83 (22.46–50.94) |
| TaqMan (n = 30) | 0.90 (0.87–0.92) | 0.80 (0.74–0.85) | 0.88 (0.82–0.93) | 6.82 (4.27–10.88) | 0.23 (0.17–0.30) | 30.15 (15.03–60.45) |
| Type of miRNA reference | ||||||
| RNU (n = 27) | 0.93 (0.91–0.95) | 0.86 (0.80–0.91) | 0.89 (0.81–0.94) | 8.08 (4.42–14.78) | 0.16 (0.11–0.23) | 51.49 (22.59–117.35) |
| Cel-miRNA (n = 50) | 0.90 (0.87–0.92) | 0.82 (0.78–0.85) | 0.85 (0.81–0.89) | 5.55 (4.22–7.29) | 0.21 (0.17–0.26) | 26.36 (17.70–39.25) |
| Included studies size | ||||||
| Sample size ≥ 100 (n = 51) | 0.89 (0.86–0.91) | 0.79 (0.75–0.83) | 0.86 (0.81–0.90) | 5.82 (4.04–8.37) | 0.24 (0.19–0.31) | 24.00 (13.91–41.42) |
| Sample size <100 (n = 51) | 0.93 (0.90–0.95) | 0.85 (0.81–0.89) | 0.87 (0.83–0.90) | 6.65 (5.03–8.78) | 0.17 (0.13–0.21) | 39.75 (27.00–58.53) |
| Different population | ||||||
| Caucasian (n = 21) | 0.95 (0.93–0.97) | 0.86 (0.78–0.91) | 0.94 (0.86–0.97) | 14.15 (5.89–34.00) | 0.15 (0.09–0.24) | 95.63 (28.08–325.74) |
| East Asian (n = 78) | 0.89 (0.86–0.91) | 0.80 (0.77–0.83) | 0.84 (0.81–0.87) | 5.12 (4.13–6.35) | 0.23 (0.20–0.28) | 21.88 (15.97–29.99) |
| Type of different miRNAs | ||||||
| miR-1 (n = 11) | 0.88 (0.85–0.90) | 0.78 (0.71–0.84) | 0.86 (0.77–0.91) | 5.41 (3.18–9.19) | 0.26 (0.18–0.36) | 21.07 (9.17–48.38) |
| miR-133 a/b (n = 9) | 0.94 (0.92–0.96) | 0.85 (0.72–0.92) | 0.92 (0.78–0.98) | 10.79 (3.63–32.09) | 0.17 (0.09–0.31) | 64.18 (19.03–216.49) |
| miR-133a (n = 5) | 0.93 (0.91–0.95) | 0.85 (0.69–0.94) | 0.92 (0.61–0.99) | 10.63 (1.69–66.96) | 0.16 (0.07–0.36) | 66.15 (7.41–590.92) |
| miR-208 a/b (n = 13) | 0.94 (0.91–0.95) | 0.83 (0.74–0.89) | 0.98 (0.88–0.99) | 35.45 (5.90–212.88) | 0.18 (0.11–0.28) | 201.13 (24.36–1660.71) |
| miR-208b (n = 7) | 0.91 (0.88–0.93) | 0.80 (0.69–0.88) | 0.96 (0.77–0.99) | 19.30 (2.78–134.22) | 0.21 (0.12–0.35) | 92.61 (9.07–945.66) |
| miR-499 (n = 17) | 0.96 (0.94–0.97) | 0.85 (0.77–0.91) | 0.95 (0.89–0.98) | 16.27 (7.31–36.22) | 0.16 (0.10–0.26) | 103.54 (31.08–345.01) |
miR, microRNA. AUC, the area under cure in the overall summary receiver operator characteristic (SROC) curve. CI, confidence interval; PLR, positive likelihood ratio. NLR, negative likelihood ratio; DOR, diagnostic odds ratio; n, the number of included studies.
Panels of Multiple miRNAs
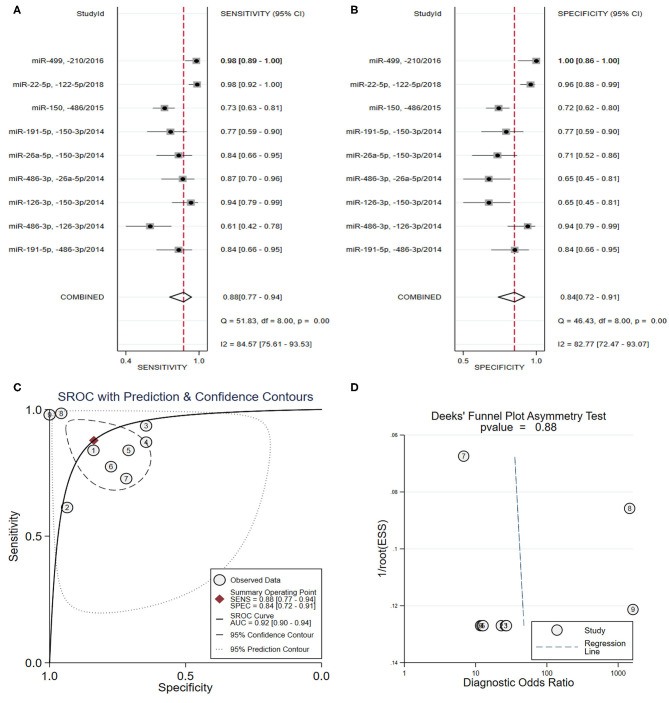
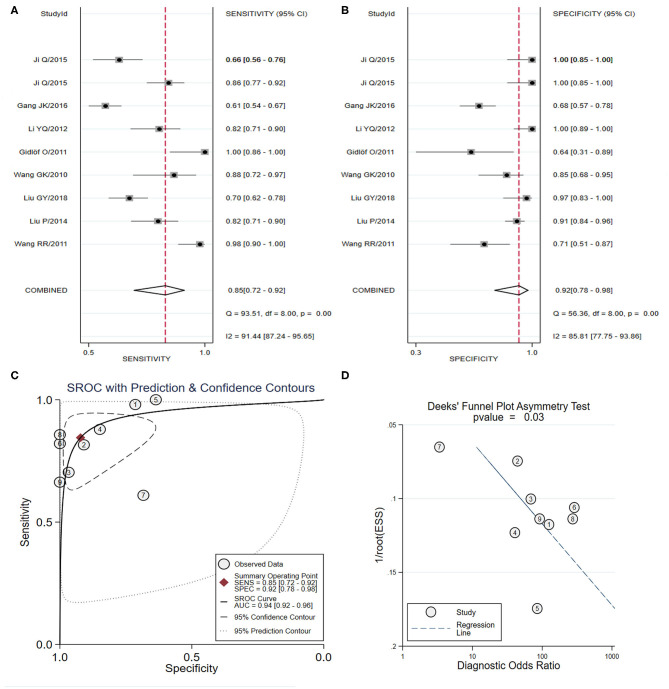
As shown in Table 3, four studies (Hsu et al., 2014; Zhang R. et al., 2015; Shalaby et al., 2016; Wang et al., 2019) focused on the diagnostic value of a panel of two types of miRNAs, and the pooled sensitivity (Figure 3A) and specificity (Figure 3B) estimates were 0.88 (95% CI: 0.77–0.94) and 0.84 (95% CI: 0.72–0.91), respectively. The area under the SROC curve (Figure 3C) was 0.92 (95% CI: 0.90–0.94). The Deeks' test (Figure 3D) was performed to evaluate publication bias, and results suggested a low probability of publication bias.
Figure 3.
The sensitivity, specificity, summary receiver operator characteristic (SROC) curve with area under curve (AUC), and funnel graph of the combination of two miRNAs in the diagnosis of acute myocardial infarction. (A) Sensitivity. (B) Specificity. (C) SROC curve with AUC. (D) Funnel graph.
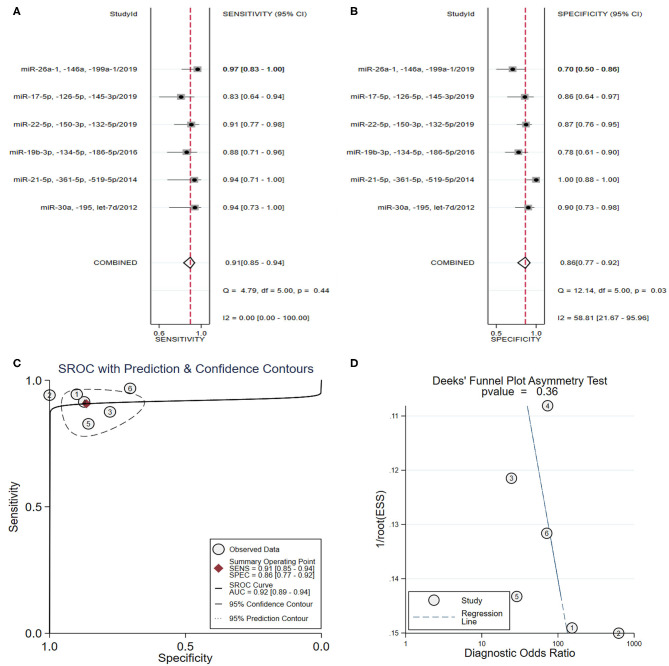
From the analysis of a panel of three types of miRNAs in six studies (Long et al., 2012b; Wang et al., 2014, 2016; Ke-Gang et al., 2016; Li H. et al., 2019; Xue et al., 2019a,b) (Table 3), the pooled sensitivity (Figure 4A) and specificity (Figure 4B) estimates were 0.91 (95% CI: 0.85–0.94) and 0.87 (95% CI: 0.77–0.92), respectively. The area under the SROC curve (Figure 4C) was 0.92 (95% CI: 0.89–0.94). The Deeks' test was performed and suggested that publication bias likely had a low effect on the summary estimates (Figure 4D).
Figure 4.
The sensitivity, specificity, summary receiver operator characteristic (SROC) curve with area under curve (AUC), and funnel graph of the combination of three miRNAs in the diagnosis of acute myocardial infarction. (A) Sensitivity. (B) Specificity. (C) SROC curve with AUC. (D) Funnel graph.
Sensitivity Analyses
Sensitivity analyses were performed on the included studies according to the following factors: (1) Type of patient blood sample (plasma vs. serum): the SROC values were 0.92 vs. 0.89, the pooled sensitivity and specificity were 0.84 vs. 0.78 and 0.87 vs. 0.87, respectively; (2) Type of miRNA detection method (SYBR green vs TaqMan): the SROC values were 0.92 vs. 0.90, the pooled sensitivity and specificity were 0.84 vs. 0.80 and 0.87 vs. 0.88, respectively; (3) Type of miRNA reference used for normalization (RNU or Cel-miRNA): the SROC values were 0.93 vs. 0.90, the pooled sensitivity and specificity were 0.86 vs. 0.82 and 0.89 vs. 0.85, respectively; (4) different study sizes (sample size ≥ 100 vs. sample size <100): the SROC values were 0.89 vs 0.93, the pooled sensitivity and specificity were 0.79 vs. 0.85 and 0.86 vs. 0.87, respectively; (5) Different populations (Caucasian vs. East Asian): the SROC values were 0.95 vs. 0.89, the pooled sensitivity and specificity were 0.86 vs. 0.80 and 0.94 vs. 0.84, respectively. A summary of the sensitivity analysis results is shown in Table 4. According to compared diagnostic values from each of the above groups, no major improvement or differences were found in the diagnostic accuracy values.
Meta-Analysis by Types of miRNAs
miRNA-1
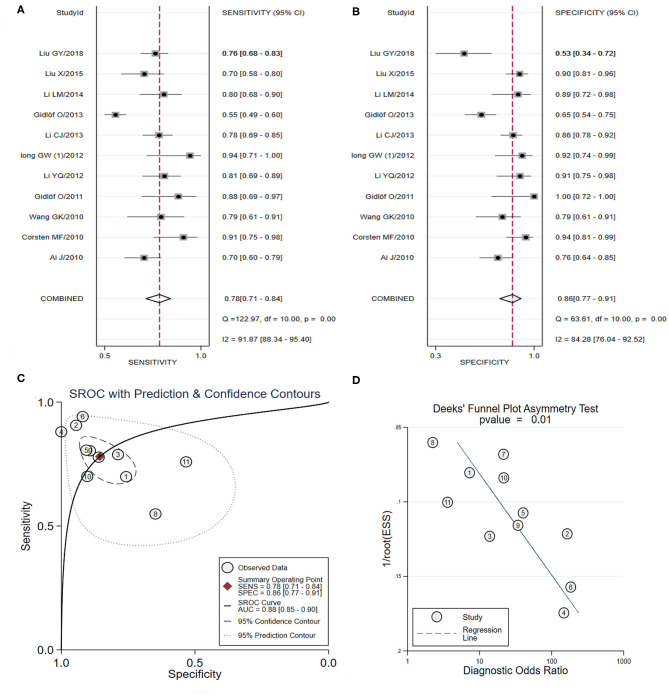
In total, 11 studies identifying the diagnostic value of miRNA-1 in AMI were included in the meta-analysis (Ai et al., 2010; Corsten et al., 2010; Wang et al., 2010; Gidlöf et al., 2011, 2013; Long et al., 2012a; Li C. et al., 2013; Li Y. Q. et al., 2013; Li L. M. et al., 2014; Liu et al., 2015, 2018) (Table 3). As shown in Figures 5A,B, the pooled sensitivity and specificity estimates were 0.78 (95% CI: 0.71–0.84) and 0.86 (95% CI: 0.77–0.91), respectively. The area under the SROC curve for miRNA-1 was 0.88 (95% CI: 0.85–0.90) (Figure 5C). The pooled PLR was 5.41 (95% CI: 3.18–9.19), and the pooled NLR was 0.26 (95% CI: 0.18–0.36). The DOR was 21.07 (95% CI: 9.17–48.38). Additionally, the Deeks' test suggested that publication bias may have some effect on the summary estimates (p-value = 0.01) (Figure 5D). Representative results from the above analyses are shown in Table 4.
Figure 5.
The sensitivity, specificity, summary receiver operator characteristic (SROC) curve with area under curve (AUC), and funnel graph of miRNA-1 in the diagnosis of acute myocardial infarction. (A) Sensitivity. (B) Specificity. (C) SROC curve with AUC. (D) Funnel graph.
miRNA-133
Nine studies that focused on the diagnostic accuracy of miRNA-133 (including miRNA-133a/b) in AMI were included in the meta-analysis (Corsten et al., 2010; D'Alessandra et al., 2010; Wang et al., 2010, 2011, 2013; Gidlöf et al., 2011; Li Y. Q. et al., 2013; Olivieri et al., 2013; Peng et al., 2014; Ji et al., 2015; Ke-Gang et al., 2016; Liu et al., 2018) (Table 3). As shown in Figures 6A,B, the pooled sensitivity and specificity estimates with 95% CI were 0.85 (95% CI: 0.72–0.92) and 0.92 (95% CI: 0.78–0.98), respectively. The area under the SROC curve for miRNA-133 was 0.94 (95% CI: 0.92–0.96) (Figure 6C). The pooled PLR was 10.79 (95% CI: 3.63–32.09) and the pooled NLR was 0.17 (95% CI: 0.09–0.31). The DOR was 64.18 (95% CI: 19.03–216.49). The Deeks' test suggested a potential publication bias (p-value = 0.01) (Figure 6D). Further subgroup analysis was conducted on miRNA-133a in the diagnosis of AMI and the SROC value, the pooled sensitivity, specificity, and DOR in the five studies (Wang et al., 2010; Gidlöf et al., 2011; Li Y. Q. et al., 2013; Ji et al., 2015; Ke-Gang et al., 2016) were 0.93 (95% CI: 0.91–0.95), 0.85 (95% CI: 0.69–0.94), 0.92 (95% CI: 0.61–0.99), and 66.15 (95% CI: 7.41–590.92). Representative results from the above analyses are shown in Table 4.
Figure 6.
The sensitivity, specificity, summary receiver operator characteristic (SROC) curve with area under curve (AUC), and funnel graph of the miRNA-133 family in the diagnosis of acute myocardial infarction. (A) Sensitivity. (B) Specificity. (C) SROC curve with AUC. (D) Funnel graph.
miRNA-208
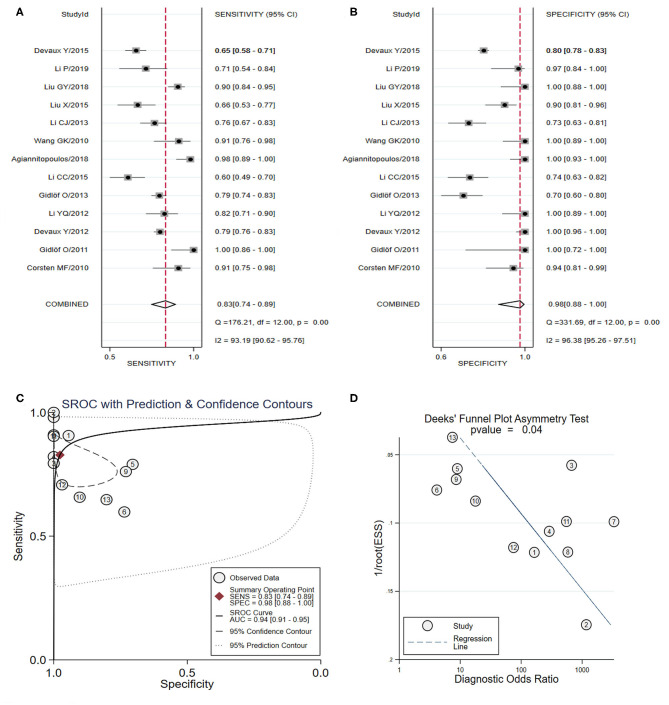
Thirteen studies evaluated the diagnostic value of miRNA-208 (including miRNA-208a/b) in AMI and were included in the meta-analysis (Corsten et al., 2010; Wang et al., 2010; Gidlöf et al., 2011, 2013; Devaux et al., 2012, 2015; Li C. et al., 2013; Li Y. Q. et al., 2013; Li et al., 2015; Liu et al., 2015, 2018; Agiannitopoulos et al., 2018; Li P. et al., 2019) (Table 3). As shown in Figures 7A,B, the pooled sensitivity and specificity estimates with 95% CI were 0.83 (95% CI: 0.74–0.89) and 0.98 (95% CI: 0.88–0.99), respectively. The area under the SROC curve for miRNA-208 was 0.94 (95% CI: 0.91–0.95) (Figure 7C). The pooled PLR was 35.45 (95% CI: 5.90–212.88), and the pooled NLR was 0.18 (95% CI: 0.11–0.28). The DOR was 201.13 (95% CI: 24.36–1660.71). The Deeks' test suggested a possible publication bias (p-value = 0.04) (Figure 7D). In the subgroup analysis of miRNA-208b in the diagnosis of AMI, the SROC value, the pooled sensitivity, specificity, and DOR in the seven studies (Corsten et al., 2010; Gidlöf et al., 2011, 2013; Devaux et al., 2012, 2015; Li Y. Q. et al., 2013; Li et al., 2015) were 0.91 (95% CI: 0.88–0.93), 0.80 (95% CI: 0.69–0.88), 0.96 (95% CI: 0.77–0.99), 92.61 (95% CI: 9.07–945.66). Representative results from the above analyses are shown in Table 4.
Figure 7.
The sensitivity, specificity, summary receiver operator characteristic (SROC) curve with area under curve (AUC), and funnel graph of the miRNA-208 family in the diagnosis of acute myocardial infarction. (A) Sensitivity. (B) Specificity. (C) SROC curve with AUC. (D) Funnel graph.
miRNA-499
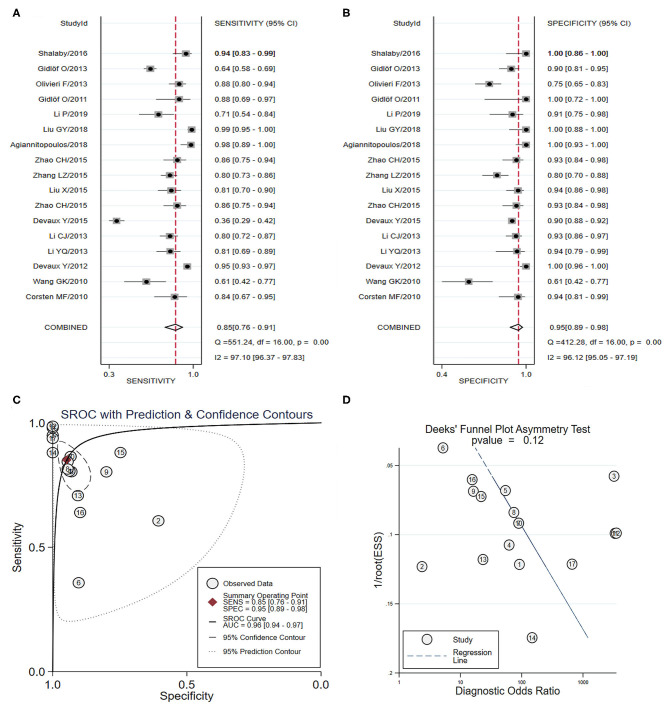
Seventeen studies involving 3,976 individuals investigated the diagnostic accuracy of miRNA-499 as a novel biomarkers for AMI (Corsten et al., 2010; Wang et al., 2010; Gidlöf et al., 2011, 2013; Devaux et al., 2012, 2015; Li C. et al., 2013; Li Y. Q. et al., 2013; Olivieri et al., 2013; Liu et al., 2015, 2018; Zhang L. et al., 2015; Zhang R. et al., 2015; Shalaby et al., 2016; Agiannitopoulos et al., 2018; Fawzy et al., 2018; Li P. et al., 2019) (Table 3). As shown in Figures 8A,B, the pooled sensitivity and specificity estimates with 95%CI were 0.85 (95% CI: 0.77–0.91) and 0.95 (95% CI: 0.89–0.98), respectively. The area under the SROC curve for miRNA-499 was 0.96 (95% CI: 0.94–0.97) (Figure 8C). The pooled PLR was 16.27 (95% CI: 7.31–36.22), and the pooled NLR was 0.16 (95% CI: 0.10–0.26). The DOR was 103.54 (95% CI: 31.08–345.01). The Deeks' test suggested a low possibility of publication bias (p-value = 0.12) (Figure 8D). Representative results from the above analyses are shown in Table 4.
Figure 8.
The sensitivity, specificity, summary receiver operator characteristic (SROC) curve with area under curve (AUC), and funnel graph of miRNA-499 in the diagnosis of acute myocardial infarction. (A) Sensitivity. (B) Specificity. (C) SROC curve with AUC. (D) Funnel graph.
Discussion
This systematic review and meta-analysis of 58 manuscripts that utilize blood circulating miRNAs (including plasma- or serum-based) in the diagnosis of AMI identified 51 significantly dysregulated miRNAs between AMI cases and controls. Additionally, this review assessed the feasibility of using these miRNAs as novel biomarkers for the diagnosis of AMI patients. Sixteen of the abnormally expressed miRNAs were investigated by more than one study, including thirteen upregulated miRNAs: miRNA-1, miRNA-19b-3p, miRNA-21, miRNA-122-5p, miRNA-126, miRNA-133a/b, miRNA-134, miRNA-150, miRNA-186, miRNA-208a/b, miRNA-486, miRNA-499, and miRNA-663b, and three downregulated miRNAs: miRNA-26a, miRNA-191, and miRNA-375. A further 34 dysregulated miRNAs were only reported by one study. The overall pooled diagnostic data of total miRNAs expression were as follows: SROC curve with AUC: 0.91, sensitivity: 0.82, specificity: 0.87, showing that circulating miRNAs might be suitable for use as potential biomarkers of AMI. Furthermore, the present meta-analysis was conducted via subgroup analyses based on type of miRNAs, including miRNA-1, miRNA-133a/b, miRNA-208a/b, and miRNA-499. MiRNA-499 had the highest diagnostic value (sensitivity: 0.85, specificity: 0.95, SROC curve with AUC: 0.96), followed by miRNA-133a (sensitivity: 0.85, specificity: 0.92, SROC curve with AUC: 0.93), and miRNA-208b had better specificity (0.96) than sensitivity (0.80). These results indicate a relatively high diagnostic accuracy for AMI based on significantly dysregulated miRNAs.
It is well-known that an early, accurate diagnosis and effective revascularization therapy play vital roles in reducing morbidity and mortality in patients with AMI (Yeh et al., 2010). At present, cardiac troponin, creatine kinase-MB (CK-MB), and myoglobin are the most widely used biomarkers in the diagnosis of AMI (de Winter et al., 1995). A good standard for the early diagnosis of AMI is an increase of cTn level (Celik et al., 2011). However, these markers are also likely to be elevated in patients with other diseases, whether or not CAD is also present (French and White, 2004). The detection of cTn has time constraints, as significant levels are reached 4–8 h following the onset of ischemia symptoms. Thus, novel genetic and molecular biomarkers of myocardial damage that have high sensitivity and specificity are still urgently needed.
Significant advances have occurred in the field of cardiovascular disease and miRNAs since their first discovery in the blood (Karakas et al., 2017). A growing number of studies indicate that the abnormal expression of miRNAs plays a critical role due to their various pathological functions in the presence of myocardial infarction (Gurha, 2016; Moghaddam et al., 2019). MiRNAs are steadily present in bodily fluids (including plasma, serum, urine, and saliva) due to protection from RNase via binding to argonaute proteins and the ability to be released from cells via microvesicles, exosomes, or bound to proteins (Meister, 2013). Moreover, recent studies have showed that cTn is more difficult to detect than miRNAs in patients with MI during the earlier acute stage, as it is usually below the cut-off value (Gidlöf et al., 2013; Zhang L. et al., 2015). This suggests a difference between these two types of biomarkers in the physiological process of myocardial infarction. Cardiac troponin releases into the blood during necrosis and during the pathological process of myocardial hypoxia and ischemia (Wu and Ford, 1999). However, miRNAs can be released in response to several forms of cellular stress occurring earlier then cell necrosis such as anoxia, lactic acidosis, and cellular edema (Edeleva and Shcherbata, 2013). Thus, experts may consider the expression levels of dysregulated miRNAs at an earlier stage of AMI, as they might be reliable candidate biomarkers for the diagnosis of AMI (Li C. et al., 2013).
In this study, the summary of single miRNAs showed that miRNA-1, miRNA-133, miRNA-208, and miRNA-499 are potential candidates for the detection of AMI, as they were most frequently detected in the previous studies. Among these individual miRNAs, circulating miRNA-499 might be an effective candidate biomarker of AMI. Some studies have demonstrated that miRNA-499 was specifically expressed in the myocardium and skeletal muscle of mammals (Xue et al., 2019a) and played a critical role in the recovery process following cardiac injury (Hosoda et al., 2011). Based on the present study, miRNA-499 has a higher sensitivity and specificity in identifying patients with AMI, and these results were similar to or better than previous studies (Cheng et al., 2014; Zhao et al., 2019). Previous studies were based on relatively small sample sizes and did not include studies published in recent years. Thus, the results of the present study are more reliable and convincing. Furthermore, we strictly considered the precise setting of specific miRNAs in the diagnosis of AMI and that assessing their diagnostic performance in combination with other biomarkers, especially highly sensitive troponin immunoassays, may be warranted. A study by Olivieri et al. showed a significant correlation between miRNA-499 and cTnT, and that the diagnosis value of this combination was superior to either one alone (Olivieri et al., 2013). However, another study reported that the diagnostic value of miRNA-499 and hs-cTnT combined was not better than either them alone (Devaux et al., 2012). Thus, due to the limited sample sizes of these studies, further research is required.
There are only a small number of high-quality studies with large sample sizes focused on the diagnostic value of miRNAs in AMI. To the best of our knowledge, this study has included the largest sample size of dysregulated miRNAs as novel biomarkers for AMI for summary and evaluation. Further high-quality studies should be performed to acquire more reliable data for use in formulating standard diagnostic criterion and to determine optimal cut-off values. Moreover, due to the different diagnostic values of miRNAs, our study demonstrated that a panel of 2 or 3 miRNAs might be superior for diagnostic accuracy. The combination of 3 miRNAs trended toward a higher sensitivity than others for use as biomarkers in the diagnosis of AMI. Therefore, to further improve the feasibility of clinical diagnosis, future research should explore the most effective combination of multiple miRNAs, especially those confirmed to have a higher utilization value in the single miRNA groups.
Aside from the many studies investigating miRNA profiles in the detection of AMI, researchers should pay closer attention to the search for the technology to detect miRNAs quickly and accurately. Methods of RNA detection tend to be time-consuming, expensive, require sophisticated techniques, and are difficult to implement for urgent testing, especially in some developing countries (Lippi et al., 2013). However, novel detection technologies developed in recent years may provide a solution to these tissues, which would also support the clinical application of miRNAs in the future. Examples of new technologies include isothermal reactions based on cleavage with DNAzyme and signal amplification, which can simultaneously amplify and detect RNA (Zhao et al., 2013). This method is thought to be immune to genomic DNA pollution and has a relatively reliable sensitivity and specificity. We believed that new, precise methods of diagnosis that can detect miRNAs very rapidly and inexpensively need to be continuously improved.
In addition, the heterogeneity of the results of this study were substantial and could not be completely resolved. We found that the sources of heterogeneity included the following: quality of included studies, age, gender proportion, regional and environmental factors, and sampling criteria. Despite the heterogeneity, we believe the results of this study are worthwhile and valuable. Results of the overall and stratified analysis of different subgroups trended toward satisfactory values of miRNAs as novel biomarkers in the diagnosis of AMI. Furthermore, it was remarkable that some publication bias was found in the present study and might imply that the potential negative results were less likely to be published.
Although this study had a satisfactory result regarding the use of miRNAs for AMI detection, conclusions should be drawn cautiously due to several limitations: (1) there was a lack of standardization due to different normalization procedures across the included studies; (2) the exclusion of non-English articles may have caused important studies to be overlooked and publication bias due to significant results being more easily published; (3) the combined analysis of multiple miRNAs for a panel were insufficient, and we were unable to conduct a meta-analysis based on the data from limited studies; and (4) the results may have been affected by the impact of inevitable clinical heterogeneity, including the general condition of the included individuals, effects of medication, and medical history.
Conclusion
The current systematic review identifies numerous miRNAs associated with AMI and suggested that miRNAs may be used as a potential biomarker for the detection of AMI. For single, stand-alone miRNAs, miRNA-499 had better diagnostic accuracy than other miRNAs. A panel of two or three miRNAs might be superior for diagnostic accuracy. To develop a diagnostic test for AMI diagnosis, we suggest that a panel of miRNAs with high sensitivity and specificity should be tested. For this purpose, large scale, high-quality studies are still required to validate the clinical application of miRNAs for AMI diagnostics, as well as to identify the precise setting of dysregulated miRNAs in patients with AMI.
Data Availability Statement
Publicly available datasets were analyzed in this study. Datasets are available through the corresponding author upon reasonable request.
Ethics Statement
All analyses were based on previous published studies; thus no ethical approval or patient consent were required. All previous published studies were approved by Ethics Committee, respectively.
Author Contributions
HC and CZ designed the study. CZ carried out the statistical analysis and participated in most of the study steps. CZ, RL, JC, KH, and MA prepared the manuscript and assisted in the study processes. YH, JZ, YZ, LW, and RZ assisted in the data collection and helped in the interpretation of the study. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank LetPub (https://www.letpub.com) for the linguistic assistance during the preparation of this manuscript. Additionally, we thank Tianjin Chest Hospital Labor Union.
Footnotes
Funding. The authors gratefully acknowledge the financial support by Tianjin Chest Hospital Labor Union (HL Cong's Model Worker Innovation Studio).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00691/full#supplementary-material
Methodological quality of studies in the meta-analysis using the Quality Assessment of Diagnostic Accuracy Studies 2 score system, including risk of bias and applicability concerns. The items were scored with “yes,” “no,” or “unsure”.
Forest plots of the total miRNAs in the diagnosis of acute myocardial infarction among the studies included in the meta-analysis. (A) Sensitivity. (B) Specificity.
Summary receiver operator characteristic (SROC) curve with area under the curve (AUC) and funnel graph of the total miRNAs in the diagnosis of acute myocardial infarction. (A) SROC curve with AUC. (B) Funnel graph.
References
- Adachi T., Nakanishi M., Otsuka Y., Nishimura K., Hirokawa G., Goto Y., et al. (2010). Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin. Chem. 56, 1183–1185. 10.1373/clinchem.2010.144121 [DOI] [PubMed] [Google Scholar]
- Agiannitopoulos K., Pavlopoulou P., Tsamis K., Bampali K., Samara P., Nasioulas G., et al. (2018). Expression of miR-208b and miR-499 in Greek patients with acute myocardial infarction. In Vivo 32, 313–318. 10.21873/invivo.11239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai J., Zhang R., Li Y., Pu J., Lu Y., Jiao J., et al. (2010). Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem. Biophys. Res. Commun. 391, 73–77. 10.1016/j.bbrc.2009.11.005 [DOI] [PubMed] [Google Scholar]
- Anand A., Shah A. S. V. S., Beshiri A., Jaffe A. S., Mills N. L. (2019). Global adoption of high-sensitivity cardiac troponins and the universal definition of myocardial infarction. Clin. Chem. 65, 484–489. 10.1373/clinchem.2018.298059 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Białek S., Górko D., Zajkowska A., Kołtowski Ł., Grabowski M., Stachurska A., et al. (2015). Release kinetics of circulating miRNA-208a in the early phase of myocardial infarction. Kardiol. Pol. 73, 613–619. 10.5603/KP.a2015.0067 [DOI] [PubMed] [Google Scholar]
- Bukauskas T., Mickus R., Cereskevicius D., Macas A. (2019). Value of serum miR-23a, miR-30d, and miR-146a biomarkers in ST-elevation myocardial infarction. Med. Sci. Monit. 25, 3925–3932. 10.12659/MSM.913743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter J. V., Pan J., Rai S. N., Galandiuk S. (2016). ROC-ing along: evaluation and interpretation of receiver operating characteristic curves. Surgery 159, 1638–1645. 10.1016/j.surg.2015.12.029 [DOI] [PubMed] [Google Scholar]
- Celik S., Giannitsis E., Wollert K. C., Schwöbel K., Lossnitzer D., Hilbel T., et al. (2011). Cardiac troponin T concentrations above the 99th percentile value as measured by a new high-sensitivity assay predict long-term prognosis in patients with acute coronary syndromes undergoing routine early invasive strategy. Clin. Res. Cardiol. 100, 1077–1085. 10.1007/s00392-011-0344-x [DOI] [PubMed] [Google Scholar]
- Chen X., Zhang L., Su T., Li H., Huang Q., Wu D., et al. (2015). Kinetics of plasma microRNA-499 expression in acute myocardial infarction. J. Thorac. Dis. 7, 890–896. 10.3978/j.issn.2072-1439.2014.11.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Wang Q., You W., Chen M., Xia J. (2014). MiRNAs as biomarkers of myocardial infarction: a meta-analysis. PLoS ONE 9:e88566. 10.1371/journal.pone.0088566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Tan N., Yang J., Liu X., Cao X., He P., et al. (2010). A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin. Sci. 119, 87–95. 10.1042/CS20090645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillet J. R., Lee I., Briggs H. A., He Y., Wang K. (2014). Issues and prospects of microRNA-based biomarkers in blood and other body fluids. Molecules 19, 6080–6105. 10.3390/molecules19056080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsten M. F., Dennert R., Jochems S., Kuznetsova T., Devaux Y., Hofstra L., et al. (2010). Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Cardiovasc. Genet. 3, 499–506. 10.1161/CIRCGENETICS.110.957415 [DOI] [PubMed] [Google Scholar]
- D'Alessandra Y., Devanna P., Limana F., Straino S., Di C. A., Brambilla P. G., et al. (2010). Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur. Heart J. 31, 2765–2773. 10.1093/eurheartj/ehq167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter R. J., Koster R. W., Sturk A., Sanders G. T. (1995). Value of myoglobin, troponin T, and CK-MBmass in ruling out an acute myocardial infarction in the emergency room. Circulation 92, 3401–3407. 10.1161/01.cir.92.12.3401 [DOI] [PubMed] [Google Scholar]
- Deeks J. J., Macaskill P., Irwig L. (2005). The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J. Clin. Epidemiol. 58, 882–893. 10.1016/j.jclinepi.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Devaux Y., Mueller M., Haaf P., Goretti E., Twerenbold R., Zangrando J., et al. (2015). Diagnostic and prognostic value of circulating microRNAs in patients with acute chest pain. J. Intern. Med. 277, 260–271. 10.1111/joim.12183 [DOI] [PubMed] [Google Scholar]
- Devaux Y., Vausort M., Goretti E., Nazarov P. V., Azuaje F., Gilson G., et al. (2012). Use of circulating microRNAs to diagnose acute myocardial infarction. Clin. Chem. 58, 559–567. 10.1373/clinchem.2011.173823 [DOI] [PubMed] [Google Scholar]
- Dohi T., Maehara A., Brener S. J., Généreux P., Gershlick A. H., Mehran R., et al. (2015). Utility of peak creatine kinase-MB measurements in predicting myocardial infarct size, left ventricular dysfunction, and outcome after first anterior wall acute myocardial infarction (from the INFUSE-AMI trial). Am. J. Cardiol. 115, 563–570. 10.1016/j.amjcard.2014.12.008 [DOI] [PubMed] [Google Scholar]
- Edeleva E. V., Shcherbata H. R. (2013). Stress-induced ECM alteration modulates cellular microRNAs that feedback to readjust the extracellular environment and cell behavior. Front. Genet. 4:305. 10.3389/fgene.2013.00305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawzy M. S., Toraih E. A., Hamed E. O., Hussein M. H., Ismail H. M. (2018). Association of MIR-499a expression and seed region variant (rs3746444) with cardiovascular disease in Egyptian patients. Acta Cardiol. 73, 131–140. 10.1080/00015385.2017.1351243 [DOI] [PubMed] [Google Scholar]
- Finsterer J., Stöllberger C., Krugluger W. (2007). Cardiac and noncardiac, particularly neuromuscular, disease with troponin-T positivity. Neth. J. Med. 65, 289–295. [PubMed] [Google Scholar]
- French J. K., White H. D. (2004). Clinical implications of the new definition of myocardial infarction. Heart 90, 99–106. 10.1136/heart.90.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabisonia K., Prosdocimo G., Aquaro G. D., Carlucci L., Zentilin L., Secco I., et al. (2019). MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 569, 418–422. 10.1038/s41586-019-1191-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Guddeti R. R., Matsuzawa Y., Liu L. P., Su L. X., Guo D., et al. (2015). Plasma levels of microRNA-145 are associated with severity of Coronary Artery Disease. PLoS ONE 10:e0123477. 10.1371/journal.pone.0123477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2013 Mortality and Causes of Death Collaborators (2015). Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 385, 117–171. 10.1016/S0140-6736(14)61682-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidlöf O., Andersson P., van der Pals J., Götberg M., Erlinge D. (2011). Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology 118, 217–226. 10.1159/000328869 [DOI] [PubMed] [Google Scholar]
- Gidlöf O., Smith J. G., Miyazu K., Gilje P., Spencer A., Blomquist S., et al. (2013). Circulating cardio-enriched microRNAs are associated with long-term prognosis following myocardial infarction. BMC Cardiovasc. Disord. 13:12. 10.1186/1471-2261-13-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. L., Guo L. L., Weng Y. Q. (2017). Implication of peripheral blood miRNA-124 in predicting acute myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 21, 1054–1059. [PubMed] [Google Scholar]
- Gurha P. (2016). MicroRNAs in cardiovascular disease. Curr. Opin. Cardiol. 31, 249–254. 10.1097/HCO.0000000000000280 [DOI] [PubMed] [Google Scholar]
- Han Z., Zhang L., Yuan L., Liu X., Chen X., Ye X., et al. (2015). Change of plasma microRNA-208 level in acute myocardial infarction patients and its clinical significance. Ann. Transl. Med. 3:307. 10.3978/j.issn.2305-5839.2015.10.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Lv P., Zhao X., Wang X., Ma X., Meng W., et al. (2014). Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Mol. Cell. Biochem. 394, 137–144. 10.1007/s11010-014-2089-0 [DOI] [PubMed] [Google Scholar]
- Hoffmeister H. M., Ehlers R., Büttcher E., Steinmetz A., Kazmaier S., Helber U., et al. (2003). Relationship between minor myocardial damage and inflammatory acute-phase reaction in acute coronary syndromes. J. Thromb. Thrombolysis 15, 33–39. 10.1023/a:1026140317777 [DOI] [PubMed] [Google Scholar]
- Hosoda T., Zheng H., Cabral-da-Silva M., Sanada F., Ide-Iwata N., Ogórek B., et al. (2011). Human cardiac stem cell differentiation is regulated by a mircrine mechanism. Circulation 123, 1287–1296. 10.1161/CIRCULATIONAHA.110.982918 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hsu A., Chen S. J., Chang Y. S., Chen H. C., Chu P. H. (2014). Systemic approach to identify serum microRNAs as potential biomarkers for acute myocardial infarction. Biomed. Res. Int. 2014:418628. 10.1155/2014/418628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Chen M., Li L., He M., Hu D., Zhang X., et al. (2014). Circulating MicroRNAs and the occurrence of acute myocardial infarction in Chinese populations. Circ. Cardiovasc. Genet. 7, 189–198. 10.1161/CIRCGENETICS.113.000294 [DOI] [PubMed] [Google Scholar]
- Hung J., Teng T. H., Finn J., Knuiman M., Briffa T., Stewart S., et al. (2013). Trends from 1996 to 2007 in incidence and mortality outcomes of heart failure after acute myocardial infarction: a population-based study of 20,812 patients with first acute myocardial infarction in Western Australia. J. Am. Heart Assoc. 2:e000172. 10.1161/JAHA.113.000172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton B., Salanti G., Caldwell D. M., Chaimani A., Schmid C. H., Cameron C., et al. (2015). The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Ann. Intern. Med. 162, 777–784. 10.7326/M14-2385 [DOI] [PubMed] [Google Scholar]
- Jakob P., Kacprowski T., Briand-Schumacher S., Heg D., Klingenberg R., Stähli B. E., et al. (2017). Profiling and validation of circulating microRNAs for cardiovascular events in patients presenting with ST-segment elevation myocardial infarction. Eur. Heart J. 38, 511–515. 10.1093/eurheartj/ehw563 [DOI] [PubMed] [Google Scholar]
- Ji Q., Jiang Q., Yan W., Li X., Zhang Y., Meng P., et al. (2015). Expression of circulating microRNAs in patients with ST segment elevation acute myocardial infarction. Minerva Cardioangiol. 63, 397–402. [PubMed] [Google Scholar]
- Karakas M., Schulte C., Appelbaum S., Ojeda F., Lackner K. J., Münzel T., et al. (2017). Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease-results from the large AtheroGene study. Eur. Heart J. 38, 516–523. 10.1093/eurheartj/ehw250 [DOI] [PubMed] [Google Scholar]
- Ke-Gang J., Zhi-Wei L., Xin Z., Jing W., Ping S., Xue-Jing H., et al. (2016). Evaluating diagnostic and prognostic value of plasma miRNA133a in acute chest pain patients undergoing coronary angiography. Medicine 95:e3412. 10.1097/MD.0000000000003412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb E. J., Hall E. M., Fahie-Wilson M. (2006). Cardiac troponins and chronic kidney disease. Kidney Int. 70, 1525–1526; author reply 1526. 10.1038/sj.ki.5001802 [DOI] [PubMed] [Google Scholar]
- Li C., Chen X., Huang J., Sun Q., Wang L. (2015). Clinical impact of circulating miR-26a, miR-191, and miR-208b in plasma of patients with acute myocardial infarction. Eur. J. Med. Res. 20:58. 10.1186/s40001-015-0148-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Fang Z., Jiang T., Zhang Q., Liu C., Zhang C., et al. (2013). Serum microRNAs profile from genome-wide serves as a fingerprint for diagnosis of acute myocardial infarction and angina pectoris. BMC Med. Genomics 6:16. 10.1186/1755-8794-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhang P., Li F., Yuan G., Wang X., Zhang A., et al. (2019). Plasma miR-22-5p, miR-132-5p, and miR-150-3p are associated with acute myocardial infarction. Biomed. Res. Int. 2019:5012648. 10.1155/2019/5012648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. M., Cai W. B., Ye Q., Liu J. M., Li X., Liao X. X. (2014). Comparison of plasma microRNA-1 and cardiac troponin T in early diagnosis of patients with acute myocardial infarction. World J. Emerg. Med. 5, 182–186. 10.5847/wjem.j.1920-8642.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Li S. Y., Liu M., Ruan J. W., Wang Z. D., Xie W. C. (2019). Value of the expression of miR-208, miR-494, miR-499 and miR-1303 in early diagnosis of acute myocardial infarction. Life Sci. 232:116547. 10.1016/j.lfs.2019.116547 [DOI] [PubMed] [Google Scholar]
- Li Y. Q., Zhang M. F., Wen H. Y., Hu C. L., Liu R., Wei H. Y., et al. (2013). Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics 68, 75–80. 10.6061/clinics/2013(01)oa12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Lu J., Luo Y., Li S., Chen M. (2014). High association between human circulating microRNA-497 and acute myocardial infarction. Sci. World J. 2014:931845. 10.1155/2014/931845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G., Mattiuzzi C., Cervellin G. (2013). Circulating microRNAs (miRs) for diagnosing acute myocardial infarction: meta-analysis of available studies. Int. J. Cardiol. 167, 277–278. 10.1016/j.ijcard.2012.09.152 [DOI] [PubMed] [Google Scholar]
- Liu G., Niu X., Meng X., Zhang Z. (2018). Sensitive miRNA markers for the detection and management of NSTEMI acute myocardial infarction patients. J. Thorac. Dis. 10, 3206–3215. 10.21037/jtd.2018.05.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Fan Z., Zhao T., Cao W., Zhang L., Li H., et al. (2015). Plasma miR-1, miR-208, miR-499 as potential predictive biomarkers for acute myocardial infarction: an independent study of Han population. Exp. Gerontol. 72, 230–238. 10.1016/j.exger.2015.10.011 [DOI] [PubMed] [Google Scholar]
- Long G., Wang F., Duan Q., Chen F., Yang S., Gong W., et al. (2012a). Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int. J. Biol. Sci. 8, 811–818. 10.7150/ijbs.4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G., Wang F., Duan Q., Yang S., Chen F., Gong W., et al. (2012b). Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS ONE 7:e50926. 10.1371/journal.pone.0050926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzano S., Rost N. S., Khan M., Li H., Batista L. M., Chutinet A., et al. (2019). Early molecular oxidative stress biomarkers of ischemic penumbra in acute stroke. Neurology 93, e1288–e1298. 10.1212/WNL.0000000000008158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Q., Liang C., He Z. Q., Fan M., Wu Z. G. (2013). Circulating miR-214 is associated with the severity of coronary artery disease. J. Geriatr. Cardiol. 10, 34–38. 10.3969/j.issn.1671-5411.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahid S. S., Hornung C. A., Minor K. S., Turina M., Galandiuk S. (2006). Systematic reviews and meta-analysis for the surgeon scientist. Br. J. Surg. 93, 1315–1324. 10.1002/bjs.5596 [DOI] [PubMed] [Google Scholar]
- Meder B., Keller A., Vogel B., Haas J., Sedaghat-Hamedani F., Kayvanpour E., et al. (2011). MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res. Cardiol. 106, 13–23. 10.1007/s00395-010-0123-2 [DOI] [PubMed] [Google Scholar]
- Meister G. (2013). Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 14, 447–459. 10.1038/nrg3462 [DOI] [PubMed] [Google Scholar]
- Moghaddam A. S., Afshari J. T., Esmaeili S. A., Saburi E., Joneidi Z., Momtazi-Borojeni A. A. (2019). Cardioprotective microRNAs: lessons from stem cell-derived exosomal microRNAs to treat cardiovascular disease. Atherosclerosis 285, 1–9. 10.1016/j.atherosclerosis.2019.03.016 [DOI] [PubMed] [Google Scholar]
- Moher D., Shamseer L., Clarke M., Ghersi D., Liberati A., Petticrew M., et al. (2015). Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 4:1. 10.1186/2046-4053-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss A. J., Hall W. J., Cannom D. S., Daubert J. P., Higgins S. L., Klein H., et al. (1996). Improved survival with an implanted defibrillator in patients with coronary disease at high risk for ventricular arrhythmia. Multicenter automatic defibrillator implantation trial investigators. N. Engl. J. Med. 335, 1933–1940. 10.1056/NEJM199612263352601 [DOI] [PubMed] [Google Scholar]
- Myhre P. L., O'Meara E., Claggett B. L., de Denus S., Jarolim P., Anand I. S., et al. (2018). Cardiac troponin I and risk of cardiac events in patients with heart failure and preserved ejection fraction. Circ. Heart Fail. 11:e005312. 10.1161/CIRCHEARTFAILURE.118.005312 [DOI] [PubMed] [Google Scholar]
- Olivieri F., Antonicelli R., Lorenzi M., D'Alessandra Y., Lazzarini R., Santini G., et al. (2013). Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int. J. Cardiol. 167, 531–536. 10.1016/j.ijcard.2012.01.075 [DOI] [PubMed] [Google Scholar]
- Peng L., Chun-guang Q., Bei-fang L., Xue-zhi D., Zi-hao W., Yun-fu L., et al. (2014). Clinical impact of circulating miR-133, miR-1291 and miR-663b in plasma of patients with acute myocardial infarction. Diagn. Pathol. 9:89. 10.1186/1746-1596-9-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S., Gupta A., Shukla V., Rani V. (2018). Functional annotation of differentially expressed fetal cardiac microRNA targets: implication for microRNA-based cardiovascular therapeutics. 3 Biotech 8:494. 10.1007/s13205-018-1520-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby S. M., El-Shal A. S., Shoukry A., Khedr M. H., Abdelraheim N. (2016). Serum miRNA-499 and miRNA-210: a potential role in early diagnosis of acute coronary syndrome. IUBMB Life 68, 673–682. 10.1002/iub.1529 [DOI] [PubMed] [Google Scholar]
- Vallabhajosyula S., Sakhuja A., Geske J. B., Kumar M., Poterucha J. T., Kashyap R., et al. (2017). Role of admission troponin-T and serial troponin-T testing in predicting outcomes in severe sepsis and septic shock. J. Am. Heart Assoc. 6:5930. 10.1161/JAHA.117.005930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Long G., Zhao C., Li H., Chaugai S., Wang Y., et al. (2013). Plasma microRNA-133a is a new marker for both acute myocardial infarction and underlying coronary artery stenosis. J. Transl. Med. 11:222. 10.1186/1479-5876-11-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Long G., Zhao C., Li H., Chaugai S., Wang Y., et al. (2014). Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS ONE 9:e105734. 10.1371/journal.pone.0105734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. K., Zhu J. Q., Zhang J. T., Li Q., Li Y., He J., et al. (2010). Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 31, 659–666. 10.1093/eurheartj/ehq013 [DOI] [PubMed] [Google Scholar]
- Wang K. J., Zhao X., Liu Y. Z., Zeng Q. T., Mao X. B., Li S. N., et al. (2016). Circulating MiR-19b-3p, MiR-134-5p and MiR-186-5p are promising novel biomarkers for early diagnosis of acute myocardial infarction. Cell. Physiol. Biochem. 38, 1015–1029. 10.1159/000443053 [DOI] [PubMed] [Google Scholar]
- Wang R., Li N., Zhang Y., Ran Y., Pu J. (2011). Circulating microRNAs are promising novel biomarkers of acute myocardial infarction. Intern. Med. 50, 1789–1795. 10.2169/internalmedicine.50.5129 [DOI] [PubMed] [Google Scholar]
- Wang Y., Chang W., Zhang Y., Zhang L., Ding H., Qi H., et al. (2019). Circulating miR-22-5p and miR-122-5p are promising novel biomarkers for diagnosis of acute myocardial infarction. J. Cell. Physiol. 234, 4778–4786. 10.1002/jcp.27274 [DOI] [PubMed] [Google Scholar]
- Whiting P. F., Rutjes A. W., Westwood M. E., Mallett S., Deeks J. J., Reitsma J. B., et al. (2011). QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 155, 529–536. 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- Wu A. H., Ford L. (1999). Release of cardiac troponin in acute coronary syndromes: ischemia or necrosis. Clin. Chim. Acta 284, 161–174. 10.1016/s0009-8981(99)00078-9 [DOI] [PubMed] [Google Scholar]
- Xiao J., Shen B., Li J., Lv D., Zhao Y., Wang F., et al. (2014). Serum microRNA-499 and microRNA-208a as biomarkers of acute myocardial infarction. Int. J. Clin. Exp. Med. 7, 136–141. [PMC free article] [PubMed] [Google Scholar]
- Xue S., Liu D., Zhu W., Su Z., Zhang L., Zhou C., et al. (2019a). Circulating MiR-17-5p, MiR-126-5p and MiR-145-3p are novel biomarkers for diagnosis of acute myocardial infarction. Front. Physiol. 10:123. 10.3389/fphys.2019.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., Zhu W., Liu D., Su Z., Zhang L., Chang Q., et al. (2019b). Circulating miR-26a-1, miR-146a and miR-199a-1 are potential candidate biomarkers for acute myocardial infarction. Mol. Med. 25:18. 10.1186/s10020-019-0086-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Fu C., Xu R., Xun Z., Zhao X., Fang R. (2017). Serum microRNA-302b: the novel biomarker for diagnosis of acute myocardial infarction. Br. J. Biomed. Sci. 74, 214–216. 10.1080/09674845.2017.1333665 [DOI] [PubMed] [Google Scholar]
- Yang S. Y., Wang Y. Q., Gao H. M., Wang B., He Q. (2016). The clinical value of circulating miR-99a in plasma of patients with acute myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 20, 5193–5197. [PubMed] [Google Scholar]
- Yao X. L., Lu X. L., Yan C. Y., Wan Q. L., Cheng G. C., Li Y. M. (2015). Circulating miR-122-5p as a potential novel biomarker for diagnosis of acute myocardial infarction. Int. J. Clin. Exp. Pathol. 8, 16014–16019. [PMC free article] [PubMed] [Google Scholar]
- Yeh R. W., Sidney S., Chandra M., Sorel M., Selby J. V., Go A. S. (2010). Population trends in the incidence and outcomes of acute myocardial infarction. N. Engl. J. Med. 362, 2155–2165. 10.1056/NEJMoa0908610 [DOI] [PubMed] [Google Scholar]
- Yi J., An Y. (2018). Circulating miR-379 as a potential novel biomarker for diagnosis of acute myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 22, 540–546. 10.26355/eurrev_201801_14207 [DOI] [PubMed] [Google Scholar]
- Zhang J., Li Y., Zhao Q. (2018). Circulating miR-23b as a novel biomarker for early risk stratification after st-elevation myocardial infarction. Med. Sci. Monit. 24, 1517–1523. 10.12659/msm.908060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen X., Su T., Li H., Huang Q., Wu D., et al. (2015). Circulating miR-499 are novel and sensitive biomarker of acute myocardial infarction. J. Thorac. Dis. 7, 303–308. 10.3978/j.issn.2072-1439.2015.02.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Lan C., Pei H., Duan G., Huang L., Li L. (2015). Expression of circulating miR-486 and miR-150 in patients with acute myocardial infarction. BMC Cardiovasc. Disord. 15:51. 10.1186/s12872-015-0042-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu Y. J., Liu T., Zhang H., Yang S. J. (2016). Plasma microRNA-21 is a potential diagnostic biomarker of acute myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 20, 323–329. [PubMed] [Google Scholar]
- Zhao C. H., Cheng G. C., He R. L., Hong Y., Wan Q. L., Wang Z. Z., et al. (2015). Analysis and clinical significance of microRNA-499 expression levels in serum of patients with acute myocardial infarction. Genet. Mol. Res. 14, 4027–4034. 10.4238/2015.April.27.17 [DOI] [PubMed] [Google Scholar]
- Zhao J., Yu H., Yan P., Zhou X., Wang Y., Yao Y. (2019). Circulating MicroRNA-499 as a diagnostic biomarker for acute myocardial infarction: a meta-analysis. Dis. Markers 2019:6121696. 10.1155/2019/6121696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhou L., Tang Z. (2013). Cleavage-based signal amplification of RNA. Nat. Commun. 4:1493. 10.1038/ncomms2492 [DOI] [PubMed] [Google Scholar]
- Zhu J., Yao K., Wang Q., Guo J., Shi H., Ma L., et al. (2016). Circulating miR-181a as a potential novel biomarker for diagnosis of acute myocardial infarction. Cell. Physiol. Biochem. 40, 1591–1602. 10.1159/000453209 [DOI] [PubMed] [Google Scholar]
- Zile M. R., Mehurg S. M., Arroyo J. E., Stroud R. E., DeSantis S. M., Spinale F. G. (2011). Relationship between the temporal profile of plasma microRNA and left ventricular remodeling in patients after myocardial infarction. Circ. Cardiovasc. Genet. 4, 614–619. 10.1161/CIRCGENETICS.111.959841 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methodological quality of studies in the meta-analysis using the Quality Assessment of Diagnostic Accuracy Studies 2 score system, including risk of bias and applicability concerns. The items were scored with “yes,” “no,” or “unsure”.
Forest plots of the total miRNAs in the diagnosis of acute myocardial infarction among the studies included in the meta-analysis. (A) Sensitivity. (B) Specificity.
Summary receiver operator characteristic (SROC) curve with area under the curve (AUC) and funnel graph of the total miRNAs in the diagnosis of acute myocardial infarction. (A) SROC curve with AUC. (B) Funnel graph.
Data Availability Statement
Publicly available datasets were analyzed in this study. Datasets are available through the corresponding author upon reasonable request.