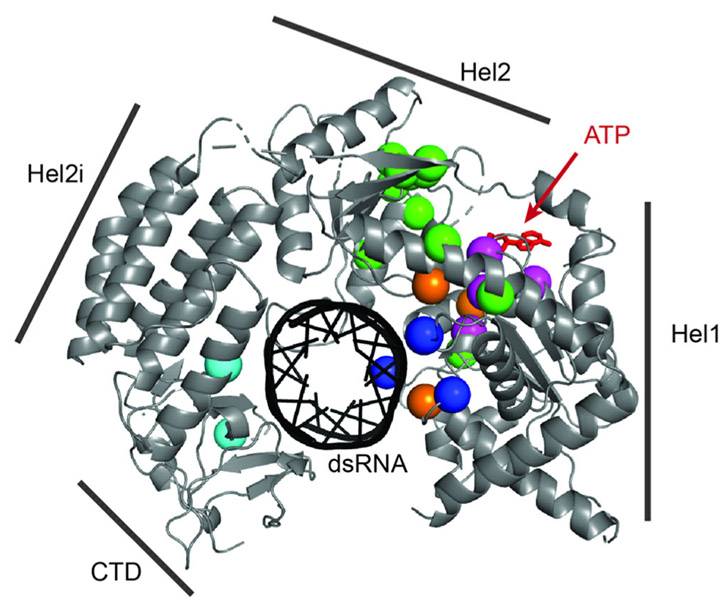
FIGURE 3.
Mutation mapping. Structure of human IFIH1 (4GL2) in complex with double stranded RNA (dsRNA; blue stick model in the center). Only the RNA binding domain (helicase domain and C-terminal domain, CTD) are included in the crystal structure. Note that the helicase domain consists of Hel1, Hel2i, and Hel2. Mutations are indicated by spheres using the following color code: residues in the ATP binding pocket (magenta), residues in the dsRNA binding surface (blue), residues within the main cluster but not directly involved in RNA binding or ATP binding (green), residues outside the main cluster (cyan), and residues previously reported by others but not in our cohort (orange). We considered all 27 mutations reported here plus three previously published mutations (p.Leu372Phe; p.Ala452Thr; p.Glu813Asp) not ascertained in our series. Residues p.Arg822, p.Arg824, and p.Ile956 are not shown because they are disordered in the crystal structure, but are expected to be located in the ATP binding (p.Arg822 and p.Arg824) and RNA binding (p.Ile956) pockets