Figure 2. LDTFs and SDTFs as contextual determinants of TGF-β–SMAD action.
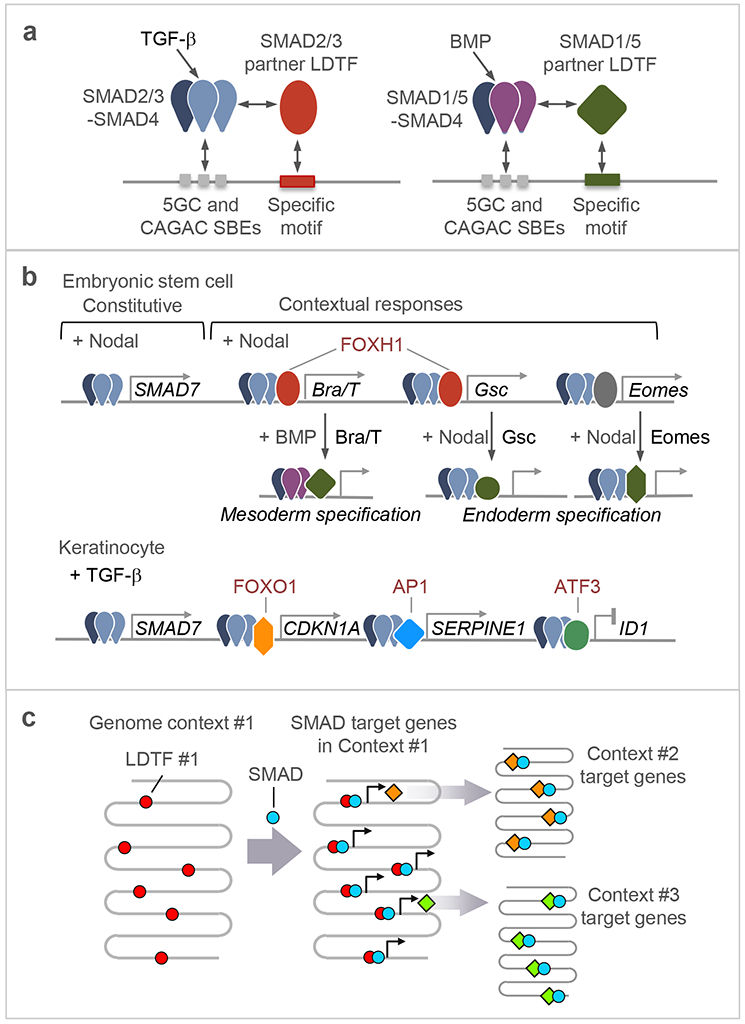
a. All R-SMADs recognize a common set of DNA elements including GGCGC and related 5bp motifs (5GC motifs), as well as the CAGAC motif. TGF-β-activated SAMAD2/3 and BMP-activated SMAD1/5 recognize different partner LDTFs, thereby achieving specificity in target gene recognition.
b. Examples of interactions between SMAD complexes and DNA binding cofactors. In ES cells poised to undergo mesendodermal differentiation, the LDTF FOXH1 is pre-bound to loci throughout the genome and recruits Nodal-activated SMAD2/3:SMAD4 complexes to these loci. The target genes encode LDTFs like BRA/T and GSC. These and the related LDTF EOMES, interact in turn with Nodal-activated or BMP-activated SMADs to mediate further specification of the mesoderm and endoderm lineages (refer also to Table 2). In keratinocyte precursors, TGF-β-activated SMAD2/3:SMAD4 complexes bind the genome in partnership with different SDTFs to regulate the expression of different subsets of target genes. Although SMADs target largely different gene sets in different cell types as a function of context-specific LDTFs and SDTFs, certain SMAD-mediated gene responses appear to be constitutive to the pathway and common to many cell types, as in the case of the negative feedback regulator SMAD7.
c. Schematic representation of successive waves of SMAD-LDTF driven gene expression programs. An LDTF that defines a particular phenotypic context (Context #1) within a given cell lineage recruits signal-driven SMADs to activate the expression of other LDTFs, which in turn interact with SMADs to mediate transitions into new differentiated states (Context #2, #3).
