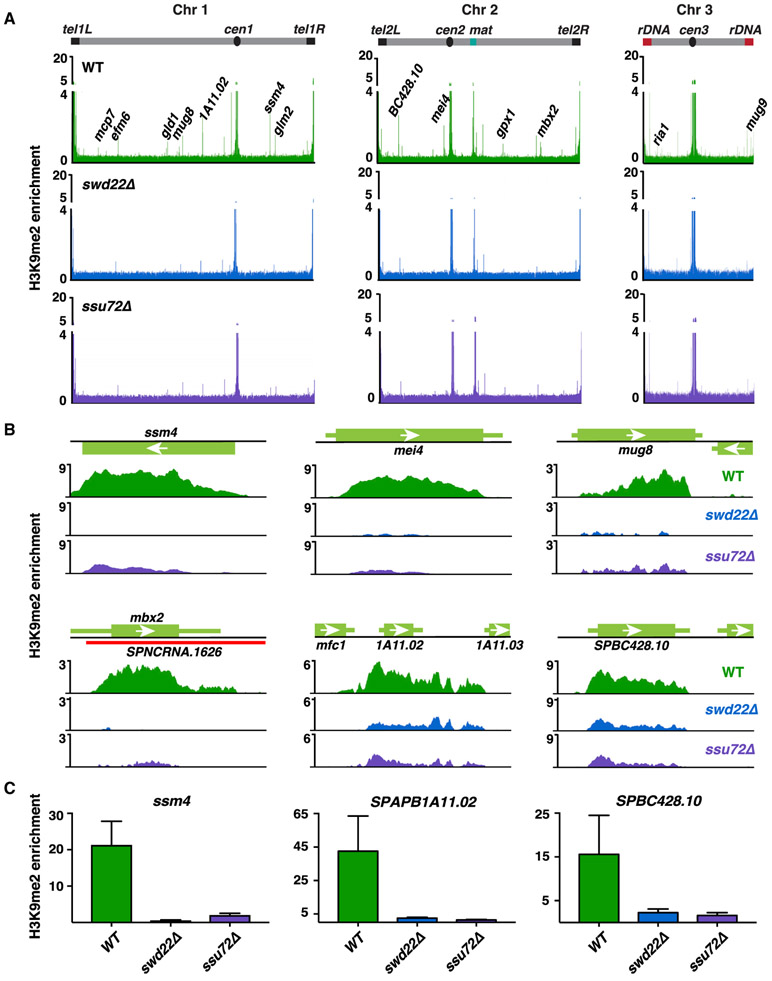
Figure 2. CPF Is Required for the Assembly of Facultative Heterochromatin Domains.
(A) Genome-wide ChIP-seq analyses of H3K9me2 in WT, swd22Δ, or ssu72Δ cells at 18°C. Representative meiotic or 18°C facultative heterochromatin islands are annotated. Enrichment values are reads per genomic content (RPGC) and represent ratios of immunoprecipitated chromatin over whole-cell extract. Islands are listed in Table S1.
(B) H3K9me2 ChIP-seq enrichment in the indicated strains at 18°C. Shown are meiotic islands at loci ssm4, mei4, mug8, and mbx2. Also shown are islands at SPAPB1A11.02 and SPBC426.10 formed in the WT at 18°C.
(C) H3K9me2 ChIP-qPCR analysis of WT, swd22Δ, or ssu72Δ cells at 18°C. Enrichment values are fold changes relative to the leu1 control. n = 3. Error bars are shown as SDs from the mean.