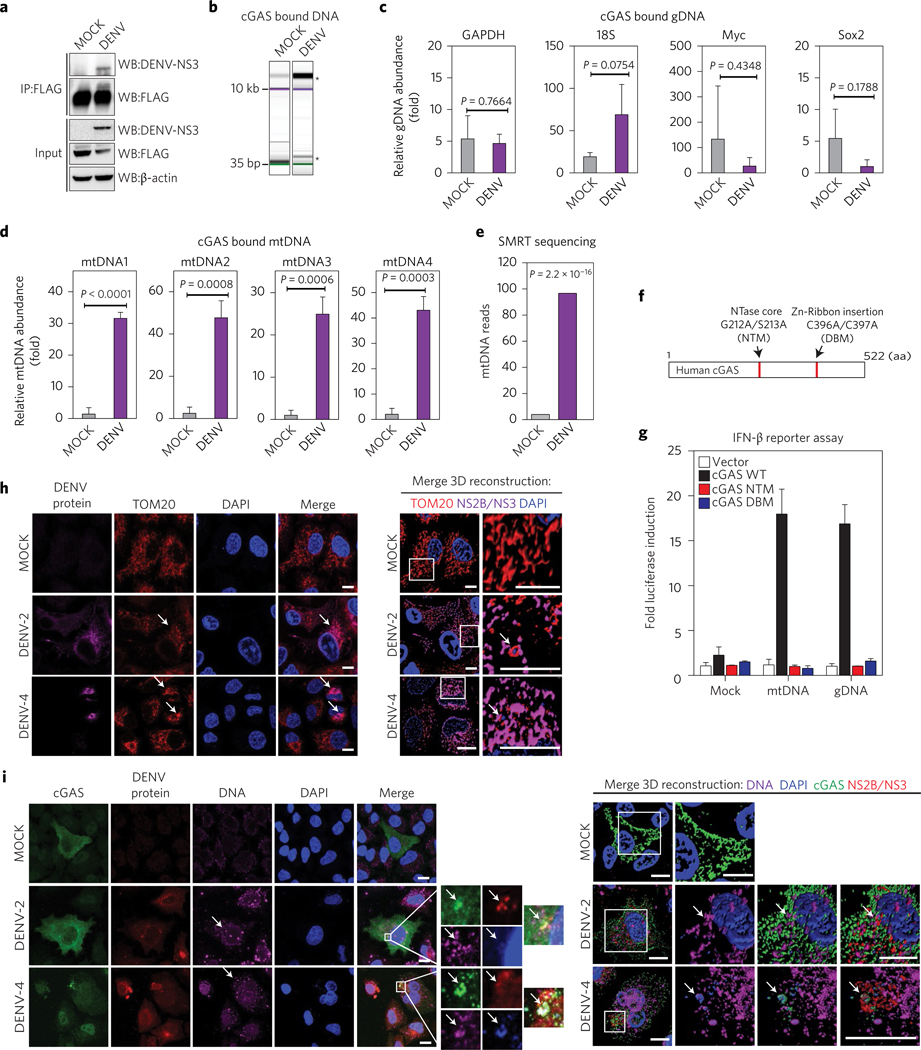
Figure 6 |. cGAS binds mitochondrial DNA (mtDNA) during DENV infection.
a, cGAS immunoprecipitation (IP). Total lysates and IP fractions were used to detect cGAS-FLAG, DENV NS3 and β-actin by SDS–PAGE/western blot. b, Analysis of DNA bound to IP cGAS from MOCK, DENV conditions by DNA electrophoresis (Bioanalyzer). *Enriched DNA bands. Relative enrichment of genomic DNA in cGAS pulldown from MOCK and DENV conditions. c, RT–qPCR for GAPDH, 18S, Myc and Sox2 DNAs, relative to EGFP DNA. d, Relative enrichment of mtDNA in cGAS pulldown from both conditions. RT–qPCR for four sets of primers that amplify fragments from different regions of the human mtDNA43. Data represent mean ± s.e.m. from three IP experiments, (unpaired Student’s t-test). e, SMRT sequencing analysis of purified DNA from cGAS pulldowns in MOCK or DENV conditions (Fisher’s exact test). f, Schematic representation of human cGAS aminoacidic sequence, NTase mutant (G212A/S213A) or DNA binding core domain mutant (C396A/C397A). NTM, nucleotidyltransferase mutant; DBM, DNA binding mutant. g, IFN-reporter assay. 293T–STING–IFN-β–Luc expressing cGAS WT, or cGAS mutants were stimulated with MOCK, mtDNA or gDNA and luciferase induction was measured. Data are expressed as mean ± s.e.m. from three biological replicates h, Analysis of mitochondria distribution and morphology during DENV infection by immunofluorescence. A549 cells were infected MOCK or with DENV-2 or DENV-4. At 24 h.p.i. cells were fixed and stained with TOM20, DENV-2 NS2B or DENV-4 NS3. Alexa Fluor-conjugated secondary antibodies −568 (red) and −647 (magenta) were used to detect the primary antibodies, respectively. Nuclei were stained with DAPI (blue). Arrows mark co-localization of aggregated mitochondria and viral proteins. A detailed three-dimensional reconstruction of the Z-stacks is shown on the right. i, Detection of cytoplasmic DNA by immunofluorescence. A549 cells expressing cGAS-V5 were infected MOCK or with DENV-2 or DENV-4. Primary antibodies against V5 (cGAS), single-stranded DNA, DENV-2 NS2B or DENV-4 NS3 protein were used. Alexa Fluor-conjugated secondary antibodies −488 (green), −647 (magenta) and −568 (red) were used to detect the primary antibodies, respectively. Nuclei were stained with DAPI (blue). Images on the right indicate the squared zoom in each panel. A detailed three-dimensional reconstruction of the Z-stacks is shown on the right. Scale bars, 10 μm. Arrows mark co-localization of cGAS with viral protein and DNA.