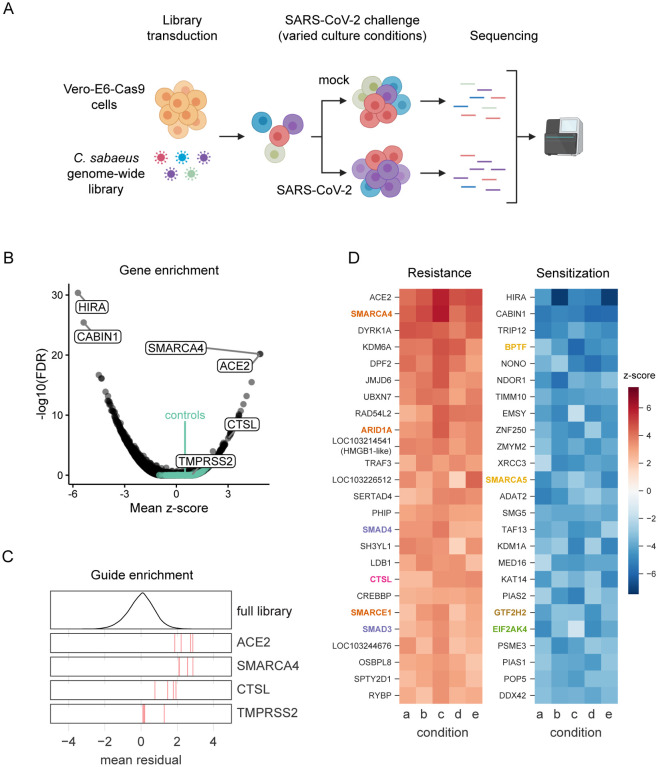
Fig. 1. Genome-wide CRISPR screen identifies genes critical for SARS-CoV-2-induced cell death.
(A) Schematic of pooled screen. Vero-E6 cells expressing Cas9 were transduced with the genome-wide C. sabaeus library via lentivirus. The transduced cell population then either received a mock treatment or was challenged with SARS-CoV-2 under various culture conditions. Surviving cells from each condition were isolated and the sgRNA sequences were amplified by PCR and sequenced. (B) Volcano plot showing top genes conferring resistance and sensitivity to SARS-CoV-2. The gene-level z-score and -log10(FDR) were both calculated using the mean of the five Cas9-v2 conditions. Non-targeting control sgRNAs were randomly grouped into sets of 4 to serve as “dummy” genes and are shown in green. (C) Performance of individual guide RNAs targeting ACE2, SMARCA4, CTSL, and TMPRSS2. The mean residual across the five Cas9-v2 conditions is plotted for the full library (top) and for the 4 guide RNAs targeting each gene. (D) Heatmaps of the top 25 gene hits for resistance and sensitivity, ranked by mean z-score in the Cas9-v2 conditions.Genes that are included in one of the gene sets labeled in (Fig 2A) are colored accordingly. Condition a: Cas9v2 D5 2.5e6 Hi-MOI; b: Cas9v2 D5 5e6 Hi-MOI; c: Cas9v2 D2 5e6 Hi-MOI; d: Cas9v2 D10 5e6 Hi-MOI; e: Cas9v2 D5 2.5e6 Lo-MOI.