Abstract
Introduction
Eosinophilic esophagitis (EoE) is a clinico-pathological diagnosis characterized by esophageal dysfunction and eosinophilic infiltration of the esophagus. Demonstration of esophageal eosinophilia (more than 15 eosinophils/hpf) in biopsy specimen obtained by esophagogastroduodenoscopy (EGD) continues to be the gold standard for diagnosis and monitoring of response to therapy. There is a growing necessity for non-invasive biomarkers that can accurately diagnose this condition and assess response to therapy. While microRNAs (miRNA) are being investigated in allergic diseases, including EoE, not many studies have explored the role of salivary miRNAs in EoE. MiR-4668-5p is a particularly interesting candidate, as it is predicted to regulate TGF-beta signaling and has not previously been identified as a target in any allergy disease. We sought to further investigate the role of miR-4668 as a biomarker to characterize and monitor response to treatment with swallowed topical glucocorticoids.
Methods
After IRB approval, twenty-two adult patients with EoE were randomly enrolled to provide a saliva sample before and after 2 months of swallowed fluticasone therapy. Differences of miRNA expression before and after treatment were analyzed by paired T-test. A significance cutoff of <0.05 was used for all analyses.
Results
Expression of miR-4668 was higher in EoE vs. non-EoE subjects. The level of miR-4668 decreased in all subjects except one, with a mean fold change 0.49 ± 0.25. There was an association between miRNA expression and number of positive aeroallergens. The miR-4668 high group had a higher number of positive aeroallergen tests, while the miR-4668 low group had a greater number of subjects with drug allergies.
Conclusions
In this study, we identified that salivary miRNAs may serve as biomarkers to characterize EoE and response to topical corticosteroids. We specifically identified miR-4668 as a novel potential biomarker, which was not previously discovered as a target in EoE or any other allergic disease.
Keywords: esophagitis, eosinophilic, EoE, miRNA, microRNA, biomarker, MiR-4668
Introduction
Eosinophilic esophagitis is a chronic immune-mediated disease first described in the early 1990s.1 It affects both children and adults and is characterized clinically by symptoms of esophageal dysfunction such as dysphagia, regurgitation and food impaction.1,2 Eosinophil-predominant inflammation on esophageal biopsy specimen is critical to the diagnosis. According to consensus recommendations originally written in 2007 and updated in 2011, absence of response to a proton pump inhibitor trial was considered crucial to the diagnosis of this condition.3 However, recently published guidelines in 2017 call this diagnostic criterion into question.1 It is now believed that clinical and histological features consistent with EoE may remit with PPI therapy, hence resolution of symptoms and inflammation with PPI trial does not exclude EoE as a diagnosis. It is recommended that at least six biopsies should be obtained from different locations, with the accepted threshold for eosinophil density for diagnosis being 15 eosinophils per high power field in esophageal mucosa.1 Histology of esophageal biopsy specimen obtained invasively via EGD continues to be necessary for diagnosis as well as monitoring of activity of this disease. Numerous non- or minimally-invasive markers are under investigation but none has been shown to be accurate so far.4 Some minimally invasive devices have shown promise but results need to be corroborated in large studies.1
MicroRNAs (miRNAs) are short, endogenous RNA molecules 19 to 25 nucleotides in length that regulate expression of target genes by posttranscriptional silencing.5 Our lab and others have investigated the role of miRNAs in allergic diseases including asthma, allergic rhinitis, atopic dermatitis and EoE, and they are considered promising candidates for biomarker development.6–10 These genetic elements are easily detectable in most body fluids and tissues. Only a few studies have explored the expression and role of microRNAs in patients with EoE. Lu et al. profiled esophageal tissue miRNA expression in patients with active EoE and those responsive to glucocorticoid treatment.11 They further compared the expression profiles with those of healthy control subjects and those with non-eosinophilic esophagitis. EoE patients had 32 differentially regulated miRNAs. MiRNA-21 and -223 were most upregulated while miRNA-375 was most downregulated. Moreover, the expression levels of these miRNAs correlated with esophageal eosinophil levels. These expression profiles were reversible in patients who responded to glucocorticoid treatment.11 Plasma analysis revealed miR-146a, miR-146b, and miR-223 as the most differentially expressed miRNAs in the plasma. Zahm et al in 2014 profiled esophageal miRNA expression in pediatric patients with active EoE and compared that to healthy control subjects.12 Five miRNAs (miR-203, miR-375, miR-21, miR-223, and miR-142-3p) were shown to be significantly altered between the groups but these changes were not reflected in the circulating RNA pool. No change in expression was noted following treatment that caused resolution of esophageal eosinophilia. Furthermore, Lu et al investigated differential miRNA expression in esophageal squamous cells after IL-13 stimulation.11 MiRNA-375 expression was inversely related to the degree of inflammation in esophageal biopsies from patients with EoE. Moreover, overexpression of miR-375 could markedly modify the immune-regulatory pathways associated with IL-13 in vitro.
Surveillance by EGD with biopsy continues to be the standard of care for assessing the response to treatment in EoE patients. The need for this invasive procedure on a recurrent basis can be challenging and justifies the urgent pursuit of noninvasive markers that correlate with disease activity, have high sensitivity and specificity and are cost-effective.4 Evaluation of miRNA profile in noninvasively obtained specimen such as plasma or saliva provides a potential option for a non-invasive biomarker to monitor disease progression and responses to therapy. Lu et al in 2012 identified miR-146a, miR-146b and miR-223 as the most abundant circulating miRNAs in plasma differentially expressed between EoE and healthy individuals.13 The levels of miR-146a and miR-223 reversed in glucocorticoid responsive EoE. Saliva is a promising alternative that yields itself to noninvasive surveillance. Swanson et al presented an abstract where they profiled salivary miRNA in a pediatric cohort and discovered 97 differentially expressed miRNAs, with miR-223 having the greatest level of expression.14 Our group previously analyzed miRNA expression in saliva samples from 15 adult patients with EoE and 17 healthy controls. We identified a set of candidate miRNA biomarkers and found increased levels of miR-570-3p, miR-3613-5p, miR-4668-5p, and miR-30a- 5p in EoE samples compared to control.15 Expression of miR-3613-5p and miR-4668-5p decreased in patients following fluticasone treatment, suggesting these two miRNAs may be candidate biomarkers for surveilling treatment response.
MiR-4668-5p was a particularly interesting candidate, as it is predicted to regulate TGF-beta signaling and has not previously been identified as a target in any allergy disease. In this study, we sought to further investigate the role of miR-4668 as a biomarker to, characterize and monitor response to treatment with swallowed topical glucocorticoids.
Subjects and Methods
Human Subject Recruitment and Study Design
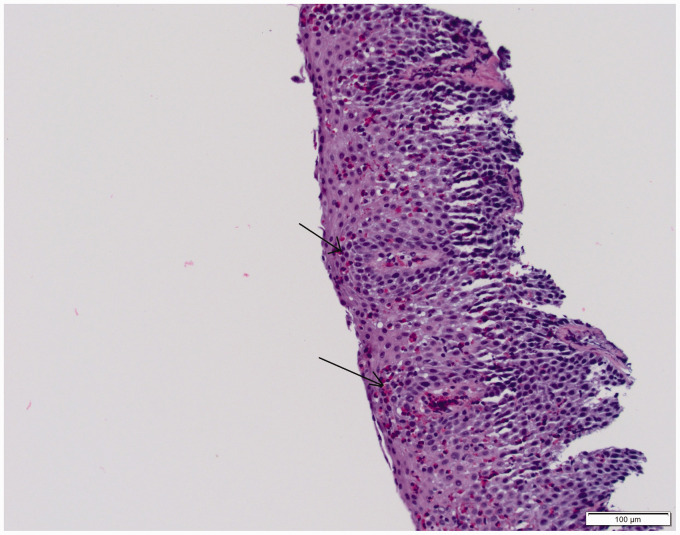
This study was approved by the Penn State College of Medicine Institutional Review Board (Study: PRAMS040665EP). In accordance with approved protocols, written informed consent was obtained from all patients. Study group comprised of 22 adults with biopsy proven diagnosis of EoE (endoscopic biopsy of esophagus, showing >15 eosinophils/hpf) and 22 subjects without EoE (Figure 1 shows a representative histopathological section). Subjects with history of autoimmune disease, HIV, cancer, congestive heart failure, diabetes mellitus, chronic liver, kidney dysfunction, pregnancy or breast feeding were excluded. Patients who agreed to participate in the study were asked to provide a sample of saliva. They were given standard of care for two months, including treatment with topical fluticasone (440 mcg swallowed twice daily through a metered dose inhaler). At the end of two months, another saliva sample was collected. This study compared expression of miRNAs in saliva before and after two months of treatment with a topical steroid in subjects with EoE. The primary outcome measure was difference in miRNA expression profile between the pre- and post-treatment groups.
Figure 1.
An H&E stained section of esophageal epithelium from a patient with EoE showing numerous intraepithelial eosinophils (arrows). Courtesy: Dr. Negar Rassaei, Assistant Professor, Penn State Health Anatomic Pathology.
Saliva Collection and Processing
Whole saliva was collected from subjects in a sterile 50 ml tube and 500 microliters was aliquoted and mixed 1.5 ml of TRIzol LS reagent (Life Technologies). RNA was isolated using the Direct-zol RNA isolation kit (Zymo Research) per manufacturer protocol and as previously reported.9 Seven µl of purified RNA was reverse transcribed using the qScript miRNA cDNA synthesis kit (Quanta bio). Expression of miRNAs was analyzed by real time PCR as described previously.9
Data Processing, Analysis, and Statistics
Differences in continuous variables was assessed by T-test or Wilcoxon rank sum, for data with normal and non-normal distribution, respectively. Differences in categorical data were determined by Fisher’s exact test. Association between miRNA expression and continuous variables was analyzed by linear regression. Expression pre- and post-therapy was analyzed by paired T-test. Data were analyzed using Prism Ver. 6.01 (Graphpad).
Results
The study population was comprised of 22 subjects with biopsy-proven diagnosis of EoE and 22 subjects without EoE. The mean age of EoE patients was 41.9 years, with a female majority (64%) and primarily white race (Table 1). Regarding atopy in EoE subjects, 77% had allergic rhinitis, 45% had asthma and 16% had atopic dermatitis. The mean maximum eosinophil count/hpf was 51.6 ± 46 among EoE subjects prior to initiation of treatment with swallowed fluticasone. The mean age of the control population was 37.8 years with a male majority of 59% and primarily white race. The incidence of allergic rhinitis and asthma was 59% and 45% respectively among controls.
Table 1.
Subject Demographics.
| Characteristics | Non-EoE Subjects (n = 22) | EoE Patients (n = 22) | P value |
|---|---|---|---|
| Age (y) | 37.8 ± 13.1 | 41.9 ± 12.9 | 0.3016 |
| Sex | |||
| Male/female, no. (%) | 13/9 (59/41) | 14/8 (64/36) | 1.000 |
| Race: White/non-white, no. (%) | 18/4 (81/19) | 21/1 (95/5) | 0.3449 |
| BMI (mean ± SD, kg/m2) | 28.8 ± 3.7 | 30.2 ± 8.9 | 0.4994 |
| Allergic rhinitis, no. (%) | 13 (59) | 17 (77) | 0.3319 |
| Asthma, no. (%) | 10 (45) | 10 (45) | 1.000 |
| Eczema, no. (%) | 2 (9) | 2 (16) | 1.000 |
| Food allergy, no. (%) | Not tested/reported | 10 (45) | N/A |
| Max. Eos/HPF | Not tested/reported | 51.6 ± 46 | N/A |
Associations between Baseline miRNA Expression and Clinical Features of EoE
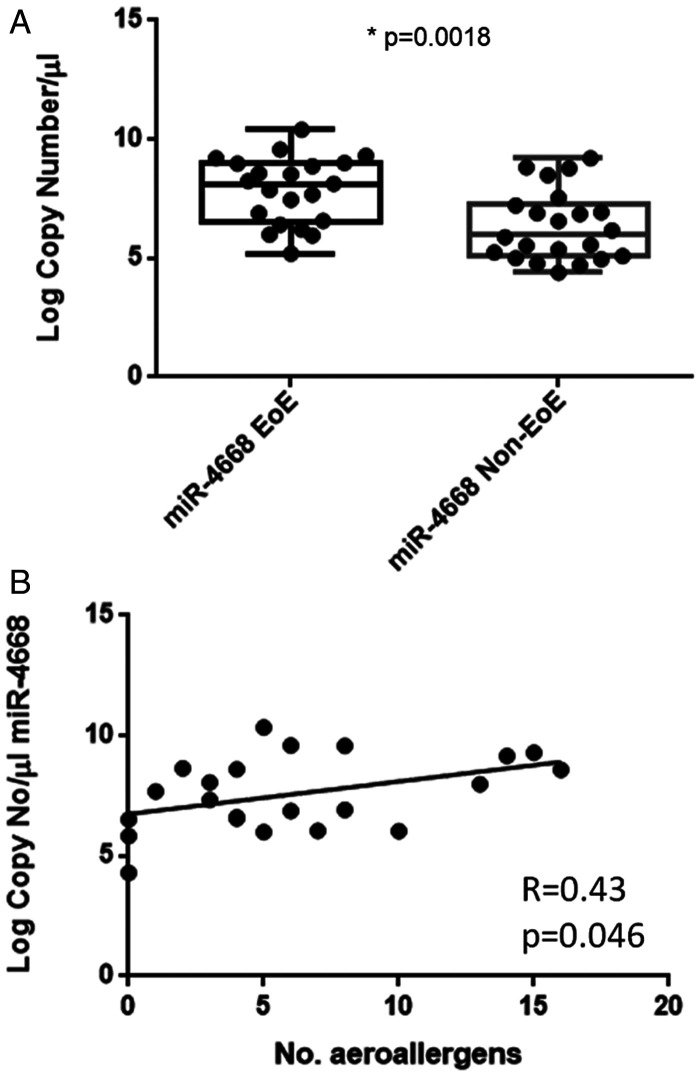
We previously utilized a qPCR array to identify a panel of candidate miRNAs whose salivary expression was different in subjects with EoE vs. those without.15 MiR-4668 emerged as a novel target, as it has not previously been studied in allergic disease. To further characterize its potential as a biomarker in EoE, we measured its expression in an expanded cohort of subjects in this study. We compared expression in whole saliva taken from EoE subjects (taken before initiation of swallowed topical steroids) to non-EoE subjects with similar atopic backgrounds. Expression of miR-4668 was higher in EoE vs. non-EoE subjects (Figure 2(A)). We next sought to determine whether its expression was associated with any clinical features of EoE and performed linear regression analysis between miR-4668 expression and continuous variables collected in the research subjects (Figure 2(B)). We found an association between miRNA expression and number of positive aeroallergen tests on a panel of 38 aeroallergens relevant to the northeast U.S. However, we did not observe any other associations between salivary miRNA expression and other clinical measures in EoE (tissue eosinophils, food allergy testing, disease severity, subject demographics such as age, sex, BMI).
Figure 2.
miR-4668 expression to characterize EoE. A, Comparison of miR-4668 expression in subjects with vs. without EoE. *p < 0.05. B, Linear regression analysis of miRNA expression vs. number of positive aeroallergen tests on a panel of relevant aeroallergens.
We next sought to determine whether miR-4668 may be used to further characterize sub-groups of patients with EoE. We categorized EoE subjects into miR-4668 “high” or miR-4668 “low” groups if expression was above or below the median miR-4668 level (Table 2). The miR-4668 high group had a higher number of positive aeroallergen tests, while the miR-4668 low group had a greater number of subjects with drug allergies. There were trends towards increased age and female sex in the miR-4668 low group, but these did not reach statistical significance.
Table 2.
Comparison of Subjects With Low and High miR-4668-5p Levels.
| miR-4668 Low (n=10) | miR-4668 High (n=12) | P Value | |
|---|---|---|---|
| Age | 37.7 ± 15.4 | 45.8 ± 10.0 | 0.15 |
| Sex: M/F, (%) | 8/10 (80%) | 6/12 (50%) | 0.204 |
| Race (W/B) | 10/0 | 11/1 | 1 |
| Positive food allergy test | 5/10 (50%) | 4/12 (33%) | 0.665 |
| Number positive aeroallergens (mean ± SD) | 9.0 ± 5.3 | 3.6 ± 2.9 | 0.0071 |
| Allergic rhinitis | 9/10 (90%) | 8/12 (67%) | 0.323 |
| Asthma | 5/10 (50%) | 5/12 (42%) | 0.665 |
| Atopic dermatitis | 1/10 (10%) | 1/12 (8%) | 1 |
| Drug allergy | 0/10 (0%) | 6/12 (50%) | 0.0152 |
| Peak tissue eosinophils/HPF (mean ± SD) | 44.1 ± 35.3 | 57.9 ± 54.0 | 0.496 |
MiRNA Expression Changes in EoE Subjects after Swallowed Topical Corticosteroids
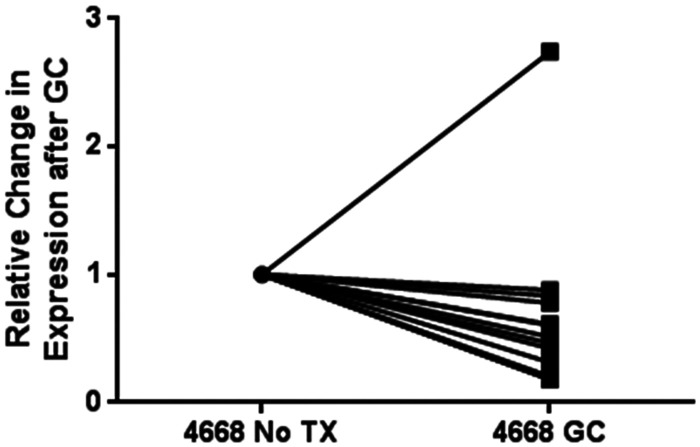
In order to determine whether therapy with swallowed topical glucocorticoids altered miRNA expression, we measured salivary levels before and after 8 weeks of fluticasone propionate HFA 220 mcg 2 puffs swallowed twice a day. The level of miR-4668 decreased in all subjects except one, with a mean fold change 0.49 ± 0.25 (Figure 3).
Figure 3.
Change in miR-4668 pre- and post-treatment with swallowed topical steroids. Relative expression change in salivary miR-4668 in EoE subjects before and after treatment with swallowed ICS.
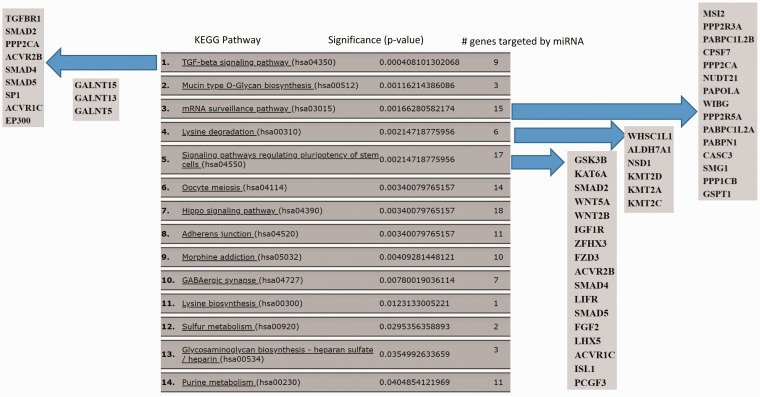
We performed an in-silico analysis to gain insight into the function of miR-4668. A miR-Path analysis (DIANA miR-Path v.3 using the microT-CDS database) identified multiple KEGG pathways regulated by the miRNA (Figure 4). The top pathway identified was TGF-β signaling, and targets included TGF-βR1, and SMADS2, 4, 5. The pathway analysis also identified tight junction regulation as a target of miR-4668, which is linked to TGF-β signaling and has been implicated in EoE.
Figure 4.
miRPath 2.0 analysis of miR-4668-5p target pathways. KEGG pathways targeted by the miRNA are shown. Individual genes targeted by miR-4668-5p shown for the top 5 pathways.
Discussion
MiRNAs are short, endogenous RNA molecules involved in post-transcriptional silencing of target genes.5,16 Extracellular miRNAs can be effectively isolated from noninvasively obtained saliva samples.14,16 Our lab as well as others have demonstrated the biomarker potential of miRNAs in many allergic diseases including asthma, atopic dermatitis and allergic rhinitis.6,8,10 In this study, we identified salivary miRNAs that may serve as biomarkers to characterize EoE and response to topical corticosteroids. We specifically identified miR-4668 as a novel potential biomarker, which was not previously discovered as a target in EoE or any other allergic disease.
Only a few studies have investigated the biomarker potential of miRNA expression in EoE. Lu et al. detected a unique miRNA signature of 32 miRNAs in esophageal tissue in EoE patients compared to non-EoE controls.13 Subsequently, Zahm et al. identified 14 miRNAs differentially expressed between EoE patients and healthy subjects.12 These two studies identified miR-375, miR-21 and miR-223 as potential diagnostic and therapeutic biomarkers for EoE. Our previous results also identified miRs-21 as a potential biomarker, suggesting that saliva may serve as a useful non-invasive source that provides similar findings as tissue biopsy. We have previously shown than miRNA expression patterns associate with different phenotypes with different severities and outcomes in allergic rhinitis and asthma.6,8,9 The finding that miR-4668 is associated with allergic sensitization in EoE indicates that miRNAs may be useful to characterize and potentially phenotype EoE. It is particularly interesting to note that the miR-4668 low group was associated with drug allergy. Associations between drug allergy and EoE has not been previously described. There could be common pathways that make patients susceptible to drug allergy and EoE, and should be an area for further study. We also saw trends towards associations of sex and age with miR-4668, but our study was not powered to characterize phenotypes of EoE. Larger studies will be needed to further investigate these findings.
The current standard of care for EoE patients continues to be periodic surveillance via biopsy specimen obtained by EGD.1 The traumatic and financial burden imposed by recurrent EGDs necessitates the quest for a biomarker that can be obtained non-invasively and that can accurately diagnose the condition as well as reflect response to therapy. We found that salivary levels of miR-4668 decreased after therapy with swallowed ICS, suggesting that this may be a candidate to assess response to therapy.
An extensive literature search did not reveal previous reports of association of miR-4668 with atopy. The finding that miR-4668 expression is associated with atopy and repressed after corticosteroid treatment raises the question about whether it plays a role in disease pathogenesis. We performed an in-silico analysis to predict function of miR-4668, as this miRNA has not been well studied, and found that targets included genes in TFG-β signaling and epithelial barrier function. Loss of epithelial barrier integrity may be important to the pathogenesis of many atopic diseases, as it allows entry of allergens and pathogens into tissue Elevated level of TGF-β1 has been observed in EoE, and a recent study demonstrated that this cytokine can disrupt barrier function by down-regulating claudin-7, a tight junction molecule.17 It remains to be determined whether miR-4668 is protective or deleterious. Predictive models identified SMADs 2,4, the TGF-β1 receptor, and ROCK2 as targets, which would suggest an inhibitory function of this miRNA on TGF-β signaling. It is possible that elevated levels of the miRNA represent a form of feedback inhibition to repress TGF-β signaling, a commonly observed feature of miRNA function.
As observed with many other miRNAs, miR-4668 is emerging as a player in not only immunity and inflammation, but in cancer as well. It has been identified as a biomarker in hepatocellular carcinoma and liposarcoma, found to be de-regulated in IgA nephropathy, and altered by mycobacteria infection of macrophages.18–21 While the exact mechanisms of miR-4668 in these diseases has not been established, its effects on CD36 may be a common link. CD36 is a plasma membrane protein that is widely distributed across many cell types and is upregulated by IL-4.22 It has been shown to be expressed on eosinophils and regulates macrophage differentiation as well as the TGF-β pathway.22 Thus, miR-4668 could regulate the TGF-β pathway at multiple levels, by altering expression of CD36 or by effects on the TGF-β1 receptor or other mediators in the signal transduction pathway.
The findings of this study show promise of miRNAs as non-invasive biomarkers to assess response to treatment in EoE. Saliva is an ideal source of biomarkers, as it can be obtained in all age groups easily. It is important to note limitations in our study. First, our sample size was relatively small, as this was a pilot study. Second, our EoE study population did not have a repeat endoscopy by the time the post-ICS treatment sample was collected. As a result, there was no histological confirmation of response to topical steroid therapy. The 8 week duration of treatment with topical steroid may not be sufficient to see a change in MiRNA and one can argue that the effect will be more prominent with longer duration of anti-inflammatory treatment. As the case with all biomarkers, larger studies will be needed to validate these findings and determine their optimal use in diagnosis and monitoring of therapy. However, this work has developed standardized methodologies that can be used for these additional studies.
Acknowledgment
We thank Theodore E. Kelbel, MD for his valuable contribution towards the compilation and analysis of data.
Ethical Approval
This study was approved by our institutional review board.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Statement of Human and Animal Rights
This article does not contain any studies with human or animal subjects.
Statement of Informed Consent
There are no human subjects in this article and informed consent is not applicable.
ORCID iD
Neeti Bhardwaj https://orcid.org/0000-0002-6043-4690
References
- 1.Lucendo AJ, Molina-Infante J, Arias Á, et al. Guidelines on eosinophilic esophagitis: evidence-based statements and recommendations for diagnosis and management in children and adults. United Eur Gastroenterol J. 2017; 5(3):335–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011; 128(1):3–20. [DOI] [PubMed] [Google Scholar]
- 3.Furuta GT, Liacouras CA, Collins MH, et al. ; First International Gastrointestinal Eosinophil Research Symposium (FIGERS) Subcommittees. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007; 133(4):1342–1363. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj N, Ghaffari G. Biomarkers for eosinophilic esophagitis: a review. Ann Allergy Asthma Immunol. 2012; 109(3):155–159. [DOI] [PubMed] [Google Scholar]
- 5.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018; 141(4):1202–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panganiban RP, Pinkerton MH, Maru SY, Jefferson SJ, Roff AN, Ishmael FT. Differential microRNA expression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol. 2012; 1(2):154–165. [PMC free article] [PubMed] [Google Scholar]
- 7.Pinkerton M, Chinchilli V, Banta E, et al. Differential expression of microRNAs in exhaled breath condensates of patients with asthma, patients with chronic obstructive pulmonary disease, and healthy adults. J Allergy Clin Immunol. 2013; 132(1):217–219. [DOI] [PubMed] [Google Scholar]
- 8.Panganiban RP, Wang Y, Howrylak J, et al. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol. 2016; 137(5):1423–1432. [DOI] [PubMed] [Google Scholar]
- 9.Panganiban RP, Lambert KA, Hsu MH, Laryea Z, Ishmael FT. Isolation and profiling of plasma microRNAs: biomarkers for asthma and allergic rhinitis. Methods. 2019; 152:48–54. [DOI] [PubMed] [Google Scholar]
- 10.Sonkoly E, Janson P, Majuri ML, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010; 126(3):581–589.e1–20. [DOI] [PubMed] [Google Scholar]
- 11.Lu TX, Lim EJ, Wen T, et al. MiR-375 is downregulated in epithelial cells after IL-13 stimulation and regulates an IL-13-induced epithelial transcriptome. Mucosal Immunol. 2012; 5(4):388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zahm AM, Menard-Katcher C, Benitez AJ, et al. Pediatric eosinophilic esophagitis is associated with changes in esophageal microRNAs. Am J Physiol Gastrointest Liver Physiol. 2014; 307(8):G803–G812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu TX, Sherrill JD, Wen T, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. 2012; 129(4):1064–1075.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swanson K, Devaraj S, Shulman R, Schaefer J, Hiremath G. Salivary microRNAs in eosinophilic esophagitis: pathobiologic implications and potential as noninvasive biomarker. Gastroenterology. 2016; 150:S673–S674. [Google Scholar]
- 15.Kelbel TE, Ghaffari G, Sena M, Ishmael FT. Salivary microRNA as a biomarker for monitoring response to treatment in eosinophilic esophagitis. J Allergy Clin Immunol. 2015; 135:AB78. [Google Scholar]
- 16.Kristen L, Punit J, Pooja J. Biomarkers and therapeutic targets: microRNA roles in the pathophysiology, diagnosis and management of eosinophilic esophagitis. J Transl Genet Genom. 2018; 2:11. [Google Scholar]
- 17.Nguyen N, Fernando SD, Biette KA, et al. TGF-β1 alters esophageal epithelial barrier function by attenuation of claudin-7 in eosinophilic esophagitis. Mucosal Immunol. 2018; 11(2):415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang N, Bu R, Duan Z, et al. Profiling and initial validation of urinary microRNAs as biomarkers in IgA nephropathy. PeerJ. 2015; 3:e990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fricke A, Cimniak AFV, Ullrich PV, et al. Whole blood miRNA expression analysis reveals miR-3613-3p as a potential biomarker for dedifferentiated liposarcoma. Cancer Biomark. 2018; 22(2):199–207. [DOI] [PubMed] [Google Scholar]
- 20.Pascut D, Krmac H, Gilardi F, et al. A comparative characterization of the circulating miRNome in whole blood and serum of HCC patients. Sci Rep. 2019; 9(1):8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das K, Saikolappan S, Dhandayuthapani S. Differential expression of miRNAs by macrophages infected with virulent and avirulent Mycobacterium tuberculosis. Tuberculosis (Edinb). 2013; 93(Suppl):S47–S50. [DOI] [PubMed] [Google Scholar]
- 22.Qin M, Wang L, Li F, et al. Oxidized LDL activated eosinophil polarize macrophage phenotype from M2 to M1 through activation of CD36 scavenger receptor. Atherosclerosis. 2017; 263:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]