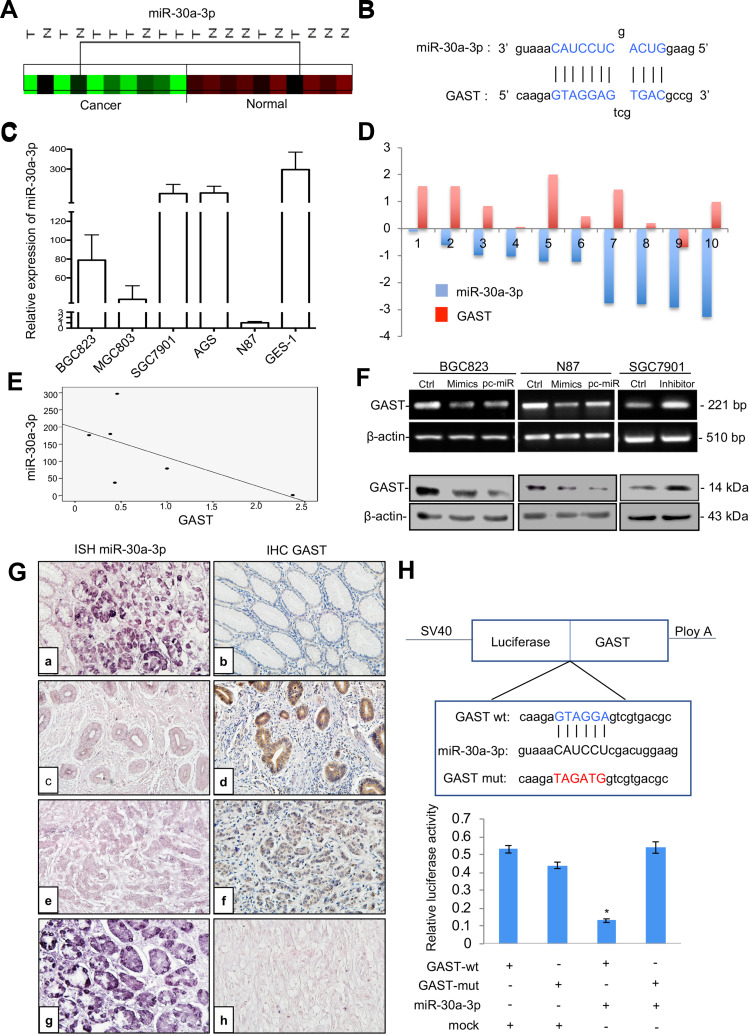
Figure 6.
GAST was the direct target gene of miR-30a-3p in GC. (A) Cluster analysis of miR-30a-3p expression patterns in our GC miRNA profiling data. (B) The binding sites of miR-30a-3p sequences in the CDS of GAST gene. (C) qPCR analysis detected miR-30a-3p expression levels in a panel of gastric cell lines. U6 was used as the endogenous control for microRNA expression analysis. (D) Histogram showing the expression of GAST and miR-30a-3p in GC tissues and matched normal tissues. The levels of GAST mRNA were quantified by RT-PCR analysis. TaqMan qPCR analysis of miR-30a-3p in 10 paired GC samples. (E) The apparently inverse correlation between GAST and miR-30a-3p expression in gastric cell lines. (F) RT-PCR (upper) and Western blots (lower) analyses were used to monitor the effect of miR-30a-3p on the expression level of GAST in BGC823 and N87 GC cells liens. (B) RT-PCR (upper) and Western blot (lower) analyses were used to monitor the effect of miR-30a-3p inhibitor or mimics on the expression level of GAST in GC cell lines, which indicated the potential inverse correlation between miR-30a-3p and GAST. (G) ISH and IHC staining assays were used to detect the expression patterns of miR-30a-3p and GAST in 31 cases of GC and matched normal ones, respectively. (A) MiR-30a-3p was highly expressed in normal tissues. (C, E) Decreased or absent miR-30a-3p expression was detected in GCs. (B) Decreased or absent GAST expression was detected in normal tissues. d, f. GAST was highly expressed in GCs. (G) DIG-labeled LNA snRNA U6 probe were used as the positive control. DIG-labeled LNA scrambled sequence probe was used as the negative control. There was a statistically significant inverse correlation between miR-30a-3p and GAST (R=−0.265, p<0.01) in these samples. (H) The results from three independent dual-luciferase reporter assays indicated that GAST was the direct targeting of miR-30a-3p. Compared with the mutant-type and wild-type of GAST CDS groups, the reporter assays showed a significantly decreased luciferase activity in the wild-type group. *p<0.01.