Figure 5: AMPK regulates HLH-30 activation and antimicrobial response upon infection with bacterial pathogens.
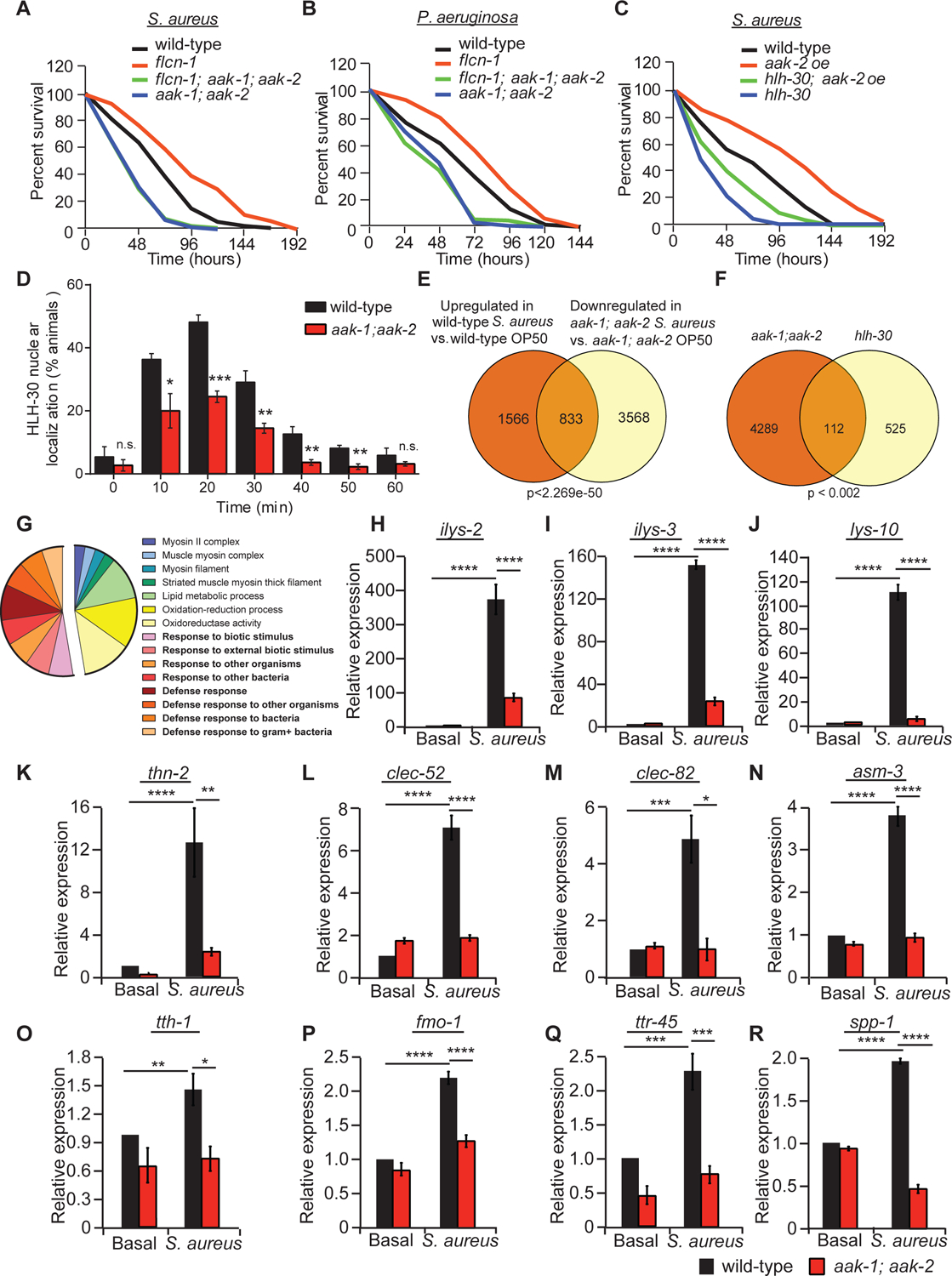
(A-C) Percent survival of indicated worm strains upon infection with S. aureus or P. aeruginosa. Refer to Table S1 for details on number of animals utilized and number of repeats. Statistics obtained by Mantel-Cox analysis on the pooled curve. (D) Percentage of animals showing HLH-30 nuclear translocation in indicated hlh-30p::hlh-30::GFP worm strains upon infection with S. aureus for the indicated amount of time. Data represent the mean ± SEM with 3 independent repeats, n ≥ 30 animals/condition for every repeat. Significance was determined using student’s t-test (*p<0.05, **p<0.01, ***p<0.001). (E) Venn diagram of the overlapping set of genes between S. aureus-induced genes in wild-type animals and genes downregulated in aak-1(tm1944); aak-2(ok524) mutant animals upon infection. (F) Venn diagram and (G) pie chart of functional gene ontology analysis of AMPK-dependent genes obtained by the overlap analysis between genes downregulated in aak-1(tm1944); aak-2(ok524) mutant animals in comparison to wild-type animals upon S. aureus infection and the hlh-30-dependent list of genes published in (40). Comparisons were performed using the “compare two lists” online software and the significance was obtained using “nemates” software. (H-R) Relative mRNA levels measured by qRT-PCR of AMPK-dependent genes in wild-type and aak-1(tm1944); aak-2(ok524) mutant animals infected with or without S. aureus for 4 h. Results are normalized to non-treated wild-type animals. Validation of RNA-seq using three biological replicates per condition and three technical replicates per biological repeat. Significance was determined using one-way ANOVA with the application of the Bonferroni correction (*p<0.05, **p<0.01, ***p<0.001, ****p<0.001).
