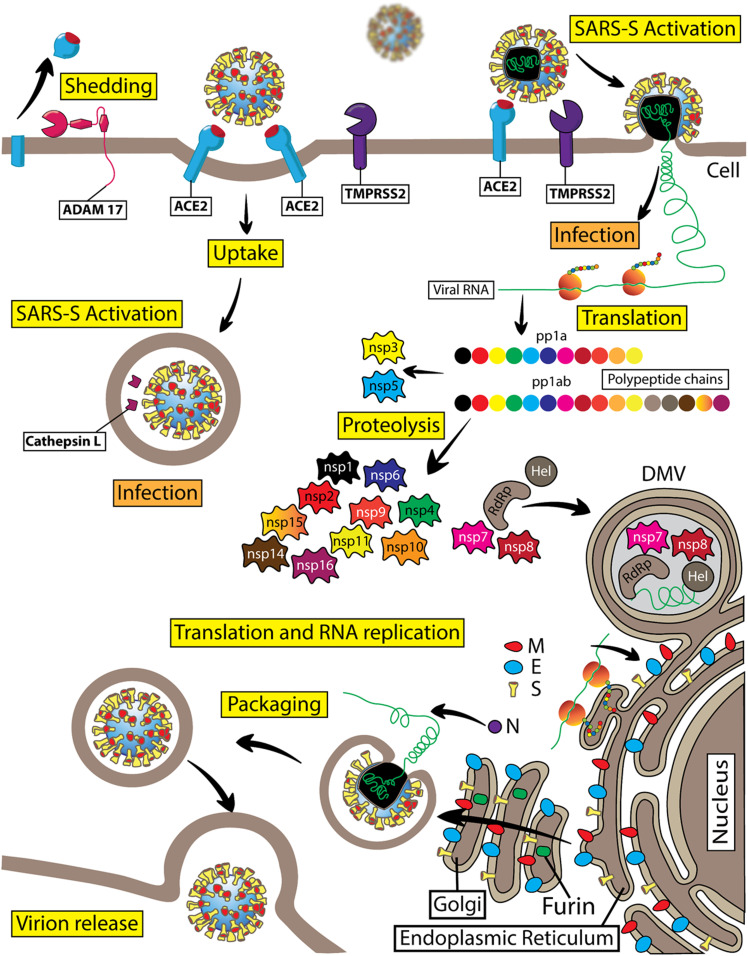
Figure 4.
Diagram of the SARS-CoV-2 virus life cycle. SARS-CoV-2 is a member of the Coronaviridae subfamily and belongs to the genus of β-coronaviruses. This is a positive-sense single-stranded RNA virus. SARS-CoV-2 viral RNA serves to code the viral genome and as mRNA for direct protein translation by the host cell ribosomes. Indeed, viral RNA contains a poly-A tail at the 3′ end and a typical mRNA cap structure at the 5′ end. SARS-CoV-2 viral RNA is nonsegmented. Viral RNA genome translation starts with the production of two replicase polyproteins, pp1a and pp1ab, which consist of 11 or 16 covalently linked nonstructural proteins (nsp), respectively. These two large polyproteins are subject to proteolytic cleavage by proteases resulting in the formation of individual nsp1–nsp16. Viral nsp3 functions as a papain-like protease and is important for cleaving the interdomain junctions between nsp1 and nsp4, whereas nsp5 is a chymotrypsin-like protease, which is also named “main protease” because it is responsible for cleaving interdomain junctions between nsp4 and nsp16. Nsp6 can induce small-diameter autophagosome formation in infected cells. Nsp12 (RdRp) is an RNA-dependent RNA polymerase, which is critical for a large-scale replication of viral RNA. Nsp12 requires several cofactors, such as nsp7 and nsp8. The RNA helicase nsp13 (Hel) is important for replication. Nsp14 is a viral N7-methyltransferase ensuring the fidelity of replication. The viral RNA also encodes four structural proteins: the spike protein (S), envelop protein (E), membrane protein (M), and nucleocapsid protein (N). In SARS-CoV-2 virions, viral RNA is enveloped with a membrane that is stabilized by the imbedded structural proteins, including S, E, M, and N proteins. The S or spike protein is a homotrimer that gives the viral particles a characteristic appearance of spiky corona. The S protein is critical for the viral entry into the host cells. The S1 subunit of the SARS-CoV-2 spike protein utilizes the hACE2 protein as its cellular receptor. The TMPRSS2 protein is the key endopeptidase that is important for priming the spike protein of SARS-CoV-2, allowing viral entry into host cells. Cathepsin B/L is an endosomal protease that can substitute TMPRSS2 activity during spike protein priming before viral RNA gains access into the cellular cytosolic compartments. SARS-CoV-2 replication takes place in double membrane vesicles (DMV) that are associated with the specific areas of the rough endoplasmic reticulum or other intracellular membranes, including autophagosomal membranes. The M, E, and N structural proteins together with the S protein are important for formation and stabilization of the SARS-CoV-2 viral particles. Viral structural protein modification takes place in the Golgi compartment before viral particles are ready for budding. Furin is a Ca2+-dependent endopeptidase enriched in the Golgi compartments that precleaves the spike protein at a specific cleavage site in the Golgi compartments, with S1 and S2 subunits remaining noncovalently bound in budding virions. ADAM17 proteolytic activity generates soluble hACE2.